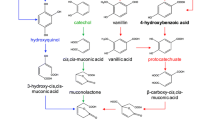
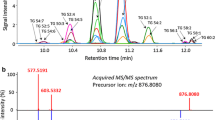
Abstract
The valorization of lignin is critical for the economic viability of the bioeconomy. Microbial metabolism is advantageous for handling the myriad of aromatic compounds resulting from lignin chemical or enzymatic depolymerization. Coupling aromatic metabolism to fatty acid biosynthesis makes possible the production of biofuels, oleochemicals, and other fine/bulk chemicals derived from lignin. Our previous work identified Cutaneotrichosporon oleaginosus as a yeast that could accumulate nearly 70% of its dry cell weight as lipids using aromatics as a sole carbon source. Expanding on this, other oleaginous yeast species were investigated for the metabolism of lignin-relevant monoaromatics. Thirty-six oleaginous yeast species from the Phaff yeast collection were screened for growth on several aromatic compounds representing S-, G-, and H- type lignin. The analysis reported in this study suggests that aromatic metabolism is largely segregated to the Cutaenotrichosporon, Trichosporon, and Rhodotorula clades. Each species tested within each clade has different properties with respect to the aromatics metabolized and the concentrations of aromatics tolerated. The combined analysis suggests that Cutaneotrichosporon yeast are the best suited to broad spectrum aromatic metabolism and support its development as a model system for aromatic metabolism in yeast.



Similar content being viewed by others
References
Davis R, Tao L, Tan ECD, Biddy M, Beckham GT, Scarlatta C, Jacobson J, Cafferty K, Ross J, Lukas J, Knorr D, Schoen P (2013) Process design and economcis for the conversion of lignocellulosic biomass to hydrocarbons: dilute-acid and enzymatic deconstruction of biomass to sugars and biological conversion of sugars to hydrocarbons. Techn Rep NREL/TP-5100–60223:88–101
DOE Bto (2016) Biorefinery optimization workshop summary report. DOE Bto, Chicago
Ibarra ML, Polk SG, Bracmort K, United States Department of Energy (2012) Biomass for biopower : feedstock supply assessments. Nova Science Publishers, New York
Holladay JE, White JF, Bozzel JJ, Johnson D (2007) Top Value-Added Chemicals from Biomass. PNNL-16983
Alinejad M, Henry C, Nikafshar S, Gondaliya A, Bagheri S, Chen N, Singh SK, Hodge DB, Nejad M (2019) Lignin-based polyurethanes: opportunities for bio-based foams, elastomers. Coat Adhes Polym (Basel). https://doi.org/10.3390/polym11071202
Kalami S, Arefmanesh M, Master E, Nejad M (2017) Replacing 100% of phenol in phenolic adhesive formulations with lignin. J Appl Polym Sci. https://doi.org/10.1002/app.45124
Gosselink RJA, de Jong E, Guran B, Abacherli A (2004) Co-ordination network for lignin - standardisation, production and applications adapted to market requirements (EUROLIGNIN). Ind Crop Prod 20:121–129. https://doi.org/10.1016/j.indcrop.2004.04.015
Jin J, Ding J, Klett A, Thies MC, Ogale AA (2018) Carbon fibers derived from fractionated-solvated lignin precursors for enhanced mechanical performance. ACS Sustain Chem Eng 6:14135–14142. https://doi.org/10.1021/acssuschemeng.8b02697
Klett AS, Chappell PV, Thies MC (2015) Recovering ultraclean lignins of controlled molecular weight from Kraft black-liquor lignins. Chem Commun 51:12855–12858. https://doi.org/10.1039/c5cc05341b
Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, Langan P, Naskar AK, Saddler JN, Tschaplinski TJ, Tuskan GA, Wyman CE (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. https://doi.org/10.1126/science.1246843
Sturgeon MR, O'Brien MH, Ciesielski PN, Katahira R, Kruger JS, Chmely SC, Hamlin J, Lawrence K, Hunsinger GB, Foust TD, Baldwin RM, Biddy MJ, Beckham GT (2014) Lignin depolymerisation by nickel supported layered-double hydroxide catalysts. Green Chem 16:824–835. https://doi.org/10.1039/c3gc42138d
Wang XY, Rinaldi R (2012) Solvent effects on the hydrogenolysis of diphenyl ether with raney nickel and their implications for the conversion of lignin. Chemsuschem 5:1455–1466. https://doi.org/10.1002/cssc.201200040
Xu CP, Arancon RAD, Labidi J, Luque R (2014) Lignin depolymerisation strategies: towards valuable chemicals and fuels. Chem Soc Rev 43:7485–7500. https://doi.org/10.1039/c4cs00235k
Beckham GT, Johnson CW, Karp EM, Salvachua D, Vardon DR (2016) Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol 42:40–53. https://doi.org/10.1016/j.copbio.2016.02.030
Henson WR, Campbell T, DeLorenzo DM, Gao Y, Berla B, Kim SJ, Foston M, Moon TS, Dantas G (2018) Multi-omic elucidation of aromatic catabolism in adaptively evolved Rhodococcus opacus. Metab Eng 49:69–83. https://doi.org/10.1016/j.ymben.2018.06.009
Lubbers RJM, Dilokpimol A, Visser J, Makela MR, Hilden KS, de Vries RP (2019) A comparison between the homocyclic aromatic metabolic pathways from plant-derived compounds by bacteria and fungi. Biotechnol Adv 37:107396. https://doi.org/10.1016/j.biotechadv.2019.05.002
Liu Z-H, Le RK, Kosa M, Yang B, Yuan J, Ragauskas AJ (2019) Identifying and creating pathways to improve biological lignin valorization. Renew Sustain Energy Rev 105:349–362. https://doi.org/10.1016/j.rser.2019.02.009
Rodriguez A, Salvachúa D, Katahira R, Black BA, Cleveland NS, Reed M, Smith H, Baidoo EEK, Keasling JD, Simmons BA, Beckham GT, Gladden JM (2017) Base-catalyzed depolymerization of solid lignin-rich streams enables microbial conversion. ACS Sustain Chem Eng 5:8171–8180. https://doi.org/10.1021/acssuschemeng.7b01818
Salvachúa D, Katahira R, Cleveland NS, Khanna P, Resch MG, Black BA, Purvine SO, Zink EM, Prieto A, Martínez MJ, Martínez AT, Simmons BA, Gladden JM, Beckham GT (2016) Lignin depolymerization by fungal secretomes and a microbial sink. Green Chem 18:6046–6062. https://doi.org/10.1039/c6gc01531j
Xie S, Sun Q, Pu Y, Lin F, Sun S, Wang X, Ragauskas AJ, Yuan JS (2017) Advanced chemical design for efficient lignin bioconversion. ACS Sustain Chem Eng 5:2215–2223. https://doi.org/10.1021/acssuschemeng.6b02401
Salvachua D, Rydzak T, Auwae R, De Capite A, Black BA, Bouvier JT, Cleveland NS, Elmore JR, Huenemann JD, Katahira R, Michener WE, Peterson DJ, Rohrer H, Vardon DR, Beckham GT, Guss AM (2019) Metabolic engineering of Pseudomonas putida for increased polyhydroxyalkanoate production from lignin. Microb Biotechnol. https://doi.org/10.1111/1751-7915.13481
Sonoki T, Takahashi K, Sugita H, Hatamura M, Azuma Y, Sato T, Suzuki S, Kamimura N, Masai E (2017) Glucose-free cis, cis-muconic acid production via new metabolic designs corresponding to the heterogeneity of lignin. ACS Sustain Chem Eng 6:1256–1264. https://doi.org/10.1021/acssuschemeng.7b03597
Vardon DR, Franden MA, Johnson CW, Karp EM, Guarnieri MT, Linger JG, Salm MJ, Strathmann TJ, Beckham GT (2015) Adipic acid production from lignin. Energy Environ Sci 8:617–628. https://doi.org/10.1039/c4ee03230f
Xu Z, Lei P, Zhai R, Wen Z, Jin M (2019) Recent advances in lignin valorization with bacterial cultures: microorganisms, metabolic pathways, and bio-products. Biotechnol Biofuels 12:32. https://doi.org/10.1186/s13068-019-1376-0
Jolivalt C, Madzak C, Brault A, Caminade E, Malosse C, Mougin C (2005) Expression of laccase IIIb from the white-rot fungus Trametes versicolor in the yeast Yarrowia lipolytica for environmental applications. Appl Microbiol Biotechnol 66:450–456. https://doi.org/10.1007/s00253-004-1717-0
Li Q, Pei J, Zhao L, Xie J, Cao F, Wang G (2014) Overexpression and characterization of laccase from Trametes versicolor in Pichia pastoris. Appl Biochem Microbiol 50:140–147. https://doi.org/10.1134/s0003683814020124
Piscitelli A, Giardina P, Mazzoni C, Sannia G (2005) Recombinant expression of Pleurotus ostreatus laccases in Kluyveromyces lactis and Saccharomyces cerevisiae. Appl Microbiol Biotechnol 69:428–439. https://doi.org/10.1007/s00253-005-0004-z
Garay LA, Sitepu IR, Cajka T, Chandra I, Shi S, Lin T, German JB, Fiehn O, Boundy-Mills KL (2016) Eighteen new oleaginous yeast species. J Ind Microbiol Biotechnol 43:887–900. https://doi.org/10.1007/s10295-016-1765-3
Sitepu I, Garay L, Sestric R, Levin D, Block DE, German J, Boundy-Mills K (2014) Oleaginous yeasts for biodiesel: Current and future trends in biology and production. J Biotechnol Adv 32:1336–1360. https://doi.org/10.1016/j.biotechadv.2014.08.003
Papanikolaou S, Aggelis G (2011) Lipids of oleaginous yeasts. Part II: technology and potential applications. Eur J Lipid Sci Technol 113:1052–1073
Probst KV, Schulte LR, Durrett TP, Rezac ME, Vadlani PV (2016) Oleaginous yeast: a value-added platform for renewable oils. Crit Rev Biotechnol 36:942–955
Alexieva Z, Gerginova M, Manasiev J, Zlateva P, Shivarova N, Krastanov A (2008) Phenol and cresol mixture degradation by the yeast Trichosporon cutaneum. J Ind Microbiol Biotechnol 35:1297–1301. https://doi.org/10.1007/s10295-008-0410-1
Anderson JJ, Dagley S (1980) Catabolism of aromatic acids in Trichosporon cutaneum. J Bacteriol 141:534–543
DeRito CM, Madsen EL (2009) Stable isotope probing reveals Trichosporon yeast to be active in situ in soil phenol metabolism. ISME J 3:477–485. https://doi.org/10.1038/ismej.2008.122
Gaal A, Neujahr H (1979) Metabolism of phenol and resorcinol in Trichosporon cutaneum. J Bacteriol 137:13–21
Powlowski JB, Dagley S (1985) β-ketoadipate pathway in Trichosporon cutaneum modified for methyl-substituted metabolites. J Bacteriol 163:1126–1135
Shoda M, Udaka S (1980) Preferential utilization of phenol rather than Glucose by Trichosporon cutaneum possessing a Partially constitutive catechol 1,2-oxygenase. Appl Environ Microbiol 39:1129–1133
Suzuki K, Itoh M (1986) Metabolism of p-hydroxybenzoate via hydroxyquinol by Trichosporon cutaneum WY2-2: characterization of the pathway using superoxide dismutase as a stabilizer of hydroxyquinol. Plant Cell Physiol 27:1451–1460
Yaguchi A, Robinson A, Mihealsick E, Blenner M (2017) Metabolism of aromatics by Trichosporon oleaginosus while remaining oleaginous. Microb Cell Fact 16:206. https://doi.org/10.1186/s12934-017-0820-8
Sànchez i Nogué V, Black BA, Kruger JS, Singer CA, Ramirez KJ, Reed ML, Cleveland NS, Singer ER, Yi X, Yeap RY, Linger JG, Beckham GT (2018) Integrated diesel production from lignocellulosic sugars via oleaginous yeast. Green Chem 20:4349–4365. https://doi.org/10.1039/c8gc01905c
Fialová A, Boschke E, Bley T (2004) Rapid monitoring of the biodegradation of phenol-like compounds by the yeast Candida maltosa using BOD measurements. Int Biodeterior Biodegrad 54:69–76. https://doi.org/10.1016/j.ibiod.2004.02.004
Gerecova G, Nebohacova M, Zeman I, Pryszcz LP, Tomaska L, Gabaldon T, Nosek J (2015) Metabolic gene clusters encoding the enzymes of two branches of the 3-oxoadipate pathway in the pathogenic yeast Candida albicans. FEMS Yeast Res. https://doi.org/10.1093/femsyr/fov006
Krug M, Ziegler H, Straube G (1985) Degradation of phenolic compounds by the yeast Candida tropicalis HP 15 I. Physiology of growth and substrate utilization. J Basic Microbiol 25:103–110
Sampaio JP (1999) Utilization of low molecular weight aromatic compounds by heterobasidiomycetous yeasts: taxonomic implications. Can J Microbiol 45:491–512
Sitepu I, Selby T, Zhu S, Lin T, Boundy-Mills K (2014) Carbon source utilization and inhibitor tolerance of 45 oleaginous yeast species. J Ind Microbiol Biotechnol 41:1061–1070. https://doi.org/10.1007/s10295-014-1447-y
Sitepu IR, Sestric R, Ignatia L, Levin D, Bruce German J, Gillies LA, Almada LA, Boundy-Mills KL (2013) Manipulation of culture conditions alters lipid content and fatty acid profiles of a wide variety of known and new oleaginous yeasts species. Biores Technol 144:360–369. https://doi.org/10.1016/j.biortech.2013.06.047
Rodriguez GM, Hussain MS, Gambill L, Gao D, Yaguchi A, Blenner M (2016) Engineering xylose utilization in Yarrowia lipolytica by understanding its cryptic xylose pathway. Biotechnol Biofuels 9:149. https://doi.org/10.1186/s13068-016-0562-6
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–469. https://doi.org/10.1093/nar/gkn180
He Z, Zhang H, Gao S, Lercher MJ, Chen WH, Hu S (2016) Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res 44:W236–241. https://doi.org/10.1093/nar/gkw370
Wang QM, Yurkov AM, Göker M, Lumbsch HT, Leavitt SD, Groenewald M, Theelen B, Liu XZ, Boekhout T, Bai FY (2015) Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Stud Mycol 81:149–189. https://doi.org/10.1016/j.simyco.2015.12.002
Gerginova M, Peneva N, Alexieva Z (2014) Investigation of feasibility of gene transfer in Trichosporon cutaneum yeast strain R57. J BioSci Biotech 2014:95–99
Görner C, Redai V, Bracharz F, Schrepfer P, Garbe D, Brück T (2016) Genetic engineering and production of modified fatty acids by the non-conventional oleaginous yeast Trichosporon oleaginosus ATCC 20509. Green Chem 18:2037–2046. https://doi.org/10.1039/c5gc01767j
Koivuranta K, Castillo S, Jouhten P, Ruohonen L, Penttila M, Wiebe MG (2018) Enhanced triacylglycerol production with genetically modified Trichosporon oleaginosus. Front Microbiol 9:1337. https://doi.org/10.3389/fmicb.2018.01337
Ochsner U, Glumoff V, Kalin M, Fiechter A, Reiser J (1991) Genetic transformation of auxotrophic mutants of the filamentous yeast Trichosporon cutaneum using homologous and heterologous marker genes. Yeast 7:513–524
Sienko M, Stepien P, Paszewski A (1992) Generation of genetic recombinants in Trichosporon cutaneum by spontaneous segregation of protoplast fusants. J Gen Microbiol 138:1409
Sampaio JP (1995) Utilization of low molecular weight lignin-related aromatic compounds for the selective isolation of yeasts: Rhodotorula vanillica, a new basidiomycetous yeast species. Syst Appl Microbiol 17:613–619. https://doi.org/10.1016/s0723-2020(11)80083-4
Mills C, Child JJ, Spencer JFT (1971) The utilization of aromatic compounds by yeasts. Antonie Van Leeuwenhoek 37:281–287
Sampio JP, Bauer R, Begerow D, Oberwinkler F (1999) Occultifur externus sp. nov., A new species of simple-pored auricularioid heterobasidiomycete from plant litter in Portugal. Mycologia 91:1094–1101
Islam MA, Yousuf A, Karim A, Pirozzi D, Khan MR, Wahid ZA (2018) Bioremediation of palm oil mill effluent and lipid production by Lipomyces starkeyi: a combined approach. J Clean Prod 172:1779–1787. https://doi.org/10.1016/j.jclepro.2017.12.012
Yousuf A, Sannino F, Addorisio V, Pirozzi D (2010) Microbial conversion of olive oil mill wastewaters into lipids suitable for biodiesel production. J Agric Food Chem 58:8630–8635. https://doi.org/10.1021/jf101282t
Juanssilfero AB, Kahar P, Amza RL, Miyamoto N, Otsuka H, Matsumoto H, Kihira C, Thontowi A, Yopi OC, Prasetya B, Kondo A (2018) Selection of oleaginous yeasts capable of high lipid accumulation during challenges from inhibitory chemical compounds. Biochem Eng J 137:182–191. https://doi.org/10.1016/j.bej.2018.05.024
Yaegashi J, Kirby J, Ito M, Sun J, Dutta T, Mirsiaghi M, Sundstrom ER, Rodriguez A, Baidoo E, Tanjore D, Pray T, Sale K, Singh S, Keasling JD, Simmons BA, Singer SW, Magnuson JK, Arkin AP, Skerker JM (2017) Gladden JM. https://doi.org/10.1101/154872
Gerginova M, Zlateva P, Peneva N, Alexieva Z (2014) Influence of phenolic substrates utilised by yeast Trichosporon cutaneum on the degradation kinetics. Biotechnol Biotechnol Equip 28:33–37. https://doi.org/10.1080/13102818.2014.901671
Chtourou M, Ammar E, Nasri M, Medhioub K (2004) Isolation of a yeast, Trichosporon cutaneum, able to use low molecular weight phenolic compounds: application to olive mill waste water treatment. J Chem Technol Biotechnol 79:869–878. https://doi.org/10.1002/jctb.1062
Wang J, Gao Q, Zhang H, Bao J (2016) Inhibitor degradation and lipid accumulation potentials of oleaginous yeast Trichosporon cutaneum using lignocellulose feedstock. Bioresour Technol 218:892–901. https://doi.org/10.1016/j.biortech.2016.06.130
Hu M, Wang J, Gao Q, Bao J (2018) Converting lignin derived phenolic aldehydes into microbial lipid by Trichosporon cutaneum. J Biotechnol 281:81–86. https://doi.org/10.1016/j.jbiotec.2018.06.341
Middelhoven W, Scorzeti G, Fell J (1999) Trichosporon guehoae sp.nov., an anamorphic basidiomycetous yeast. Can J Microbiol 45:686–690
Deeba F, Pruthi V, Negi YS (2018) Aromatic hydrocarbon biodegradation activates neutral lipid biosynthesis in oleaginous yeast. Bioresour Technol 255:273–280. https://doi.org/10.1016/j.biortech.2018.01.096
Su X, Zhou M, Hu P, Xiao Y, Wang Z, Mei R, Hashmi MZ, Lin H, Chen J, Sun F (2019) Whole-genome sequencing of an acidophilic Rhodotorula sp. ZM1 and its phenol-degrading capability under acidic conditions. Chemosphere 232:76–86. https://doi.org/10.1016/j.chemosphere.2019.05.195
Jarboui R, Baati H, Fetoui F, Gargouri A, Gharsallah N, Ammar E (2012) Yeast performance in wastewater treatment: case study of Rhodotorula mucilaginosa. Environ Technol 33:951–960. https://doi.org/10.1080/09593330.2011.603753
Rodriguez A, Ersig N, Geiselman GM, Seibel K, Simmons BA, Magnuson JK, Eudes A, Gladden JM (2019) Conversion of depolymerized sugars and aromatics from engineered feedstocks by two oleaginous red yeasts. Bioresour Technol 286:121365. https://doi.org/10.1016/j.biortech.2019.121365
Huang C, Chen X-f, Xiong L, Yang X-y, Chen X-d, Ma L-l, Chen Y (2013) Microbial oil production from corncob acid hydrolysate by oleaginous yeast Trichosporon coremiiforme. Biomass Bioenerg 49:273–278. https://doi.org/10.1016/j.biombioe.2012.12.023
Patel A, Sartaj K, Arora N, Pruthi V, Pruthi PA (2017) Biodegradation of phenol via meta cleavage pathway triggers de novo TAG biosynthesis pathway in oleaginous yeast. J Hazard Mater 340:47–56. https://doi.org/10.1016/j.jhazmat.2017.07.013
Pyne ME, Narcross L, Melgar M, Kevvai K, Mookerjee S, Leite GB, Martin VJJ (2018) An Engineered Aro1 protein degradation approach for increased cis, cis-muconic acid biosynthesis in Saccharomyces cerevisiae. Appl Environ Microbiol. https://doi.org/10.1128/AEM.01095-18
Xie NZ, Liang H, Huang RB, Xu P (2014) Biotechnological production of muconic acid: current status and future prospects. Biotechnol Adv 32:615–622. https://doi.org/10.1016/j.biotechadv.2014.04.001
Spagnuolo M, Yaguchi A, Blenner M (2019) Oleaginous yeast for biofuel and oleochemical production. Curr Opin Biotechnol 57:73–81. https://doi.org/10.1016/j.copbio.2019.02.011
Yaguchi A, Rives D, Blenner M (2017) New kids on the block: emerging oleaginous yeast of biotechnological importance. AIMS Microbiol 3:227–247. https://doi.org/10.3934/microbiol.2017.2.227
Yaguchi A, Spagnuolo M, Blenner M (2018) Engineering yeast for utilization of alternative feedstocks. Curr Opin Biotechnol 53:122–129. https://doi.org/10.1016/j.copbio.2017.12.003
McCluskey K, Barker K, Barton H, Boundy-Mills K, Brown D, Coddington J, Cook K, Desmeth P, Geiser D, Glaeser J (2017) The US Culture Collection Network responding to the requirements of the Nagoya Protocol on Access and Benefit Sharing. mBio 8:e00982–e1017
McCluskey K, Boundy-Mills K, Beattie GA (2018) Complying with the nagoya protocol to the convention on biological diversity. SIMB News 68:8–9
McCluskey K (2017) A Review of living collections with special emphasis on sustainability and its impact on research across multiple disciplines. Biopreserv Biobank 15:20–30. https://doi.org/10.1089/bio.2016.0066
Smith D, McCluskey K, Stackebrandt E (2014) Investment into the future of microbial resources: culture collection funding models and BRC business plans for biological resource centres. SpringerPlus 3:1–12
Acknowledgements
This study was funded in part by a USDA Sun Grant (2014-38502-22598) and Creative Inquiry funds from Clemson University. ALY has been supported by a GAANN Fellowship (P200A180076) from the US Department of Education. This material is based upon work supported by the US Department of Agriculture, National Institute of Food Agriculture under Award No. 2018-67009-27901. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the US Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yaguchi, A., Franaszek, N., O’Neill, K. et al. Identification of oleaginous yeasts that metabolize aromatic compounds. J Ind Microbiol Biotechnol 47, 801–813 (2020). https://doi.org/10.1007/s10295-020-02269-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-020-02269-5