- 1Department of Microbiology, Haukeland University Hospital, Helse Bergen, Bergen, Norway
- 2Department of Clinical Science, University of Bergen, Bergen, Norway
- 3Broegelmann Research Laboratory, University of Bergen, Bergen, Norway
- 4Department of Immunology and Transfusion Medicine, Haukeland University Hospital, Helse Bergen, Bergen, Norway
- 5Norway Centre for Cancer Biomarkers, University of Bergen, Bergen, Norway
The transcription factor β-catenin is able to induce tolerogenic/anti-inflammatory features in different types of dendritic cells (DCs). Monocyte-derived dendritic cells (moDCs) have been widely used in dendritic cell-based cancer therapy, but so far with limited clinical efficacy. We wanted to investigate the hypothesis that aberrant differentiation or induction of dual pro- and anti-inflammatory features may be β-catenin dependent in moDCs. β-catenin was detectable in both immature and lipopolysaccharide (LPS)-stimulated DCs. The β-catenin inhibitor ICG-001 dose-dependently increased the pro-inflammatory signature cytokine IL-12p70 and decreased the anti-inflammatory signature molecule IL-10. The β-catenin activator 6-bromoindirubin-3′-oxime (6-BIO) dose-dependently increased total and nuclear β-catenin, and this was associated with decreased IL-12p70, increased IL-10, and reduced surface expression of activation markers, such as CD80 and CD86, and increased expression of inhibitory markers, such as PD-L1. 6-BIO and ICG-001 competed dose-dependently regarding these features. Genome-wide mRNA expression analyses further underscored the dual development of pro- and anti-inflammatory features of LPS-matured moDCs and suggest a role for β-catenin inhibition in production of more potent therapeutic moDCs.
Introduction
Dendritic cells (DCs) are the most efficient antigen-presenting cells of the immune system and play a vital role in initiating the adaptive immune response and maintaining tolerance to self-antigens (1). Immature DCs continually search their environment for antigens, while mature DCs migrate to the lymph nodes and present processed antigens on their major histocompatibility (MHC) molecules to T cells. Traditionally, three different types of DCs have been considered in peripheral blood, plasmacytoid DCs (pDC) and classical or conventional DC type 1 (cDC1) and type 2 (cDC2) (2–4). Recent high-resolution technologies have revealed additional types of human blood DCs and progenitors (5, 6). Altogether, DCs comprise less than 1% of circulating blood leukocytes, and for this reason, most DC-based therapies have relied on DCs generated in vitro from the more plentiful blood monocytes [reviewed in (2–4, 7, 8)]. Monocyte-derived DCs (moDCs) are able to activate the immune system, but it may be anticipated that there is a considerable potential for the generation of more potent and robust DCs for cancer therapy.
Depending on their phenotype and type of secreted cytokines, DCs may exert either pro-inflammatory or tolerogenic function as their response to newly encountered antigens. The transcription factor β-catenin can be activated to stimulate tolerogenic features of DCs, such as cytokine, surface marker, and metabolic profiles (9–12). Surface markers associated with pro-inflammatory activation include CD80 and CD86, whereas PD-L1 and PD-L2 are considered inhibitory or tolerogenic markers (7). Interleukin 12 (IL-12p70) represents a pro-inflammatory cytokine (13) and interleukin 10 (IL-10) an anti-inflammatory or tolerogenic cytokine (14) that can be secreted from mature DCs.
Inhibiting β-catenin signaling could have a dual effect in cancer therapy, as this pathway promotes tolerogenic features of the local dendritic cells and is often activated in cancer and cancer stem cells. In the present study, β-catenin activation was achieved using a specific inhibitor of the β-catenin destruction complex, 6-bromoindirubin-3′-oxime (6-BIO) that has been found to increase β-catenin in the cell nucleus of different cell types (15). In this way the central, final part of β-catenin signaling downstream of the destruction complex can be investigated. This approach has experimental advantages because β-catenin activation is impacted by different up-stream pathways with complicated cross-talks (16). Likewise, central β-catenin inhibition was attempted using the small molecule ICG-001 that binds CREB-binding protein (CBP) to disrupt its interaction with β-catenin and inhibits CBP function as a co-activator of β-catenin-mediated transcription at regulatory genomic elements (17).
In the present study, moDCs derived from buffy coats of healthy donors were investigated and revealed the potential of mature moDCs to co-develop both pro-inflammatory and tolerogenic features assayed by IL-12p70 and IL-10 secretion, DC surface markers, and whole-genome mRNA quantification.
Materials and Methods
Generation of Monocyte-Derived Dendritic Cells
Buffy coats of healthy blood donors at the Blood bank of Haukeland University Hospital, Bergen, were used to generate human monocyte-derived dendritic cells (moDCs). Informed consents were obtained from all donors, and samples were anonymized according to the approval by the Regional Ethical Committee (#64205). Healthy donors were above 23 years of age. Peripheral blood mononuclear cells (PBMCs) were isolated by gradient centrifugation using Lymphoprep™ (Cat. No. 1114545; Axis-Shield). Pan Monocyte Isolation Kit (Cat. No. 130-096-537; MiltenyiBiotec) with the addition of CD61 MicroBeads (Cat. No. 130-051-101; MiltenyiBiotec) and LS columns (Cat. No. 130-042-401; MiltenyiBiotec) were used to separate untouched monocytes from PBMCs by indirect magnetic labeling. Monocytes were then cultured in CellGenix GMP DC medium (Cat. No. 20801-0500; CellGenix) supplemented with 20 ng/ml of IL-4 (Cat. No. 11340047; Immunotools) and 100 ng/ml of GM-CSF (Cat. No. 11343128; Immunotools) at cell densities of 1.5 × 106 per 3 ml/well in six-well plates or 0.75 × 106 per 1.5 ml/well in 12-well plates for 4 days. IL-4 and GM-CSF were replenished on day 3. The fourth-day cultures were treated with compounds at different concentrations, i.e., 6-bromoindirubin-3-oxime (6-BIO; Cat. No. S7198; Selleckchem) 1 nM to 2 μM and/or ICG-001 (Cat. No. S2662; Selleckchem) 0.5 to 8 μM (for 24 h), and 1 h later with 30 ng/ml of LPS (for 23 h). As controls, the vehicle DMSO was added in LPS-treated and un-treated (iDC) populations. The moDCs were harvested on day 5.
Western Blots
Western blots were performed as previously described (18). moDCs were lysed in RIPA-buffer (Cat. No. ab156034; Abcam) supplemented with 1:100 Protease Inhibitor Cocktail Set I (Cat. No. 535142; Calbiochem). The protein concentration was quantified using Direct Detect® Infrared Spectrometer (EMD Millipore) using Direct Detect® Assay-free Cards (Cat. No. DDAC00010-GR; Millipore). Twenty micrograms was used for each sample loaded onto BoltTM Bis-Tris Plus Gels (Cat. No. NW04120BOX; Novex; Life Technologies). The proteins were separated by SDS electrophoresis and blotted on Amersham Hybond P 0.45 PVDF blotting membrane (Cat. No. 10600069; GE Healthcare). The primary antibodies used were anti-β-catenin (Cat. No. 16051; Abcam) and anti-GAPDH (Cat. No. MAI-16757 Invitrogen). The horseradish peroxidase (HRP)-conjugated secondary antibodies used were anti-rabbit (dilution 1:2,000; Cat. No. NA934; Amersham) and anti-mouse (dilution 1:2,000; Cat. No. 170-501; Bio-RAD). SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Cat. No. 34580; Thermo Scientific) was used for visualization with Chemidoc XRS, and images were captured using Quantity One 4.6.5 software (Bio-Rad). MagicMark™ XP Western Protein Standard (Cat. No. LC5602; Invitrogen) was used as molecular weight marker. ImageJ 1.50i software (National Institutes of Health, Bethesda, MD, USA) was used to quantify the band intensity of each protein followed by normalization to its corresponding GAPDH control.
Indirect Immunofluorescence Assay
Indirect immunofluorescence assays were performed as previously described (19). The primary antibody used was anti-β-catenin (Cat. No. 16051; Abcam) at 1 μg/ml dilution, and FITC-conjugated Pierce™ goat anti-rabbit IgG (H + L) secondary antibody (Cat. No. 31635; Thermo Scientific) was used at 1:50 dilution. Cells grown on coverslips were mounted on glass slides in SlowFade™ Gold Antifade Mountant w/DAPI (Cat. No. S36939; Invitrogen). The images were captured on Leica TCS SP8 STED 3× confocal microscope using Leica Application Suite X 2.0.2.15022 software (Leica Microsystems).
Enzyme-Linked Immunosorbent Assay (ELISA)
Secretion of IL-12p70 and IL-10 in the supernatant of moDC cultures were measured using IL-12p70 Human Uncoated ELISA Kit (Cat. No. 88-7126-88; Invitrogen) and IL-10 Human Uncoated ELISA Kit (Cat. No. 88-7106-88; Invitrogen), respectively. The absorbance was measured with Synergy H1 Hybrid Multi-Mode Reader and analyzed using Gen5 2.00.18 software (BioTek). Data are presented as fold change to DMSO-treated control sample, as absolute levels of IL-10 and IL-12 secretion varied considerably between the donors.
Flow Cytometry
The phenotype of the generated moDC populations was determined by flow cytometry as described previously (20). In short, 1 × 105 moDCs were incubated with FcR-blocking reagent (Cat. No. 130-059-901; MiltenyiBiotec) before titrated amounts of a panel of nine antibodies were added for 10 min at room temperature in the dark. The antibodies used were as follows: CD83 PE-CF594 (Cat. No. 562631; BD Biosciences), HLA-DR Horizon V500 (Cat. No. 561224; BD Biosciences), CD80 Brilliant Violet 605 (Cat. No. 305225; Biolegend), CCR7 Brilliant Violet 421 (Cat. No. 353208; Biolegend), CD86 Alexa Fluor 647 (Cat. No. 305416; Biolegend), CD274 PE-Cyanine7 (Cat. No. 46-5983-42; eBioscience), CD273 PerCP-Fluor 710 (Cat. No. 46-5888-42), CD14 FITC (Cat. No. 21620143; Immunotools), and CD1a PE (Cat. No. 21270014; Immunotools). The cells were analyzed on LSR Fortessa (BD Biosciences), and further analysis was performed using FlowJo V10 software (FlowJo, LLC). Unstained samples were used to set the gates, and 1% false-positive events were accepted throughout the analysis. For each experiment, a minimum of 5,000 single events were recorded.
Luminex Microbead Cytokine Assay
The cell-free supernatants collected from the MLR cultures were thawed and measured for cytokines using Human Magnetic 25-Plex Panel (Cat. No. LHC009M; Invitrogen) according to the manufacturer's instructions. We measured and analyzed seven cytokines including interferon-α (IFN-α), interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), interleukin (IL)-6, IL-2R, IL-10, and IL-12. Luminex plates were read using the Luminex 100 System (Luminex Corporation, Austin, TX, USA) following the manufacturer's instructions. STarStation V3.0 (Build: 3810.0; Applied Cytometry Systems, Sheffield, UK) was used to analyze the data.
Mixed Leukocyte Reaction (MLR)
Allogeneic MLR was performed as previously described (20). In short, 2 × 105 monocyte-depleted allogeneic PBMCs were labeled with CFSE using Vybrant™ CFDA SE Cell Tracer Kit (Cat. No. V12883; Invitrogen) and co-cultured with 5 × 104 moDCs of different donors for 5 days in X-Vivo 20 medium (Cat. No. 04-448Q; Lonza) supplemented with 50 U/ml of IL-2 (Cat. No. 11340023; Immunotools) and 10 ng/ml of IL-7 (Cat. No. 11340073; Immunotools). The cells were harvested and analyzed on Accuri C6 flow cytometer (BD Biosciences). For each experiment, a minimum of 20,000 single events were recorded.
DNA Microarray Analyses
Genome-wide transcription profiling using Agilent microarrays has been described previously (21). Total RNA was isolated and tested for RNA integrity by 1% agarose gel electrophoresis, then converted to Cy3-labeled cRNA targets and hybridized to Agilent Whole Human Genome 44k Microarrays (Cat. No. G4845A; Agilent Technologies). Raw data were imported and analyzed in J-Express software (http://www.molmine.com) (22). We used mean spot signals as intensity measure, normalized the expression data over the entire arrays, and log2-transformed and considered genes changed more than 1.5-fold with FDR value <5% as differentially expressed genes. DNA microarray data have been deposited into the ArrayExpress database under accession number E-MTAB-8330.
RNA Sequencing and Analyses
All experiments were conducted at QIAGEN Genomic Services. The library preparation was done using TruSeq® Stranded mRNA Sample preparation kit (Illumina Inc.). The starting material (500 ng) of total RNA was mRNA enriched using the oligodT bead system. The isolated mRNA was subsequently enzymatically fragmented. Then first-strand synthesis and second-strand synthesis were performed, and the double-stranded cDNA was purified (AMPure XP, Beckman Coulter). The cDNA was end repaired, 3′ adenylated and Illumina sequencing adaptors ligated onto the fragments ends, and the library was purified (AMPure XP). The mRNA stranded libraries were pre-amplified with PCR and purified (AMPure XP). The libraries' size distribution was validated and quality inspected on a Bioanalyzer 2100 or BioAnalyzer 4200tape Station (Agilent Technologies). High-quality libraries were pooled based in equimolar concentrations based on the Bioanalyzer Smear Analysis tool (Agilent Technologies). The library pool(s) were quantified using qPCR, and the optimal concentration of the library pool was used to generate the clusters on the surface of a flow cell before sequencing on a NextSeq 500 instrument (75 cycles) according to the manufacturer instructions (Illumina Inc.).
Software Tools Used for RNA-Seq Analysis
NGS data analysis pipeline was based on the Tuxedo software package, which is a combination of open-source software, and implements peer-reviewed statistical methods. In addition, specialized software developed internally at QIAGEN Genomic Services was employed to interpret and improve the readability of the final results. The components of NGS data analysis pipeline for RNA-seq include Bowtie2 (v. 2.2.2), see (23), Tophat (v2.0.11), see (24, 25), and Cufflinks (v2.2.1), see (26, 27).
To guide the assembly process, an existing transcript annotation was used (RABT assembly). In addition, fragment bias correction was used to correct for sequence bias during library preparation (28). When comparing groups, Cuffdiff was used to calculate the FPKM (number of fragments per kilobase of transcript per million mapped fragments) and test for differential expression.
Statistical Analysis
All data were analyzed using GraphPad Prism 8 (GraphPad software). Statistical significance of the difference was calculated using one-way analysis of variance (ANOVA) with Dunnett's multiple comparisons test or two-way ANOVA with Tukey's multiple comparisons test, and 95% confidence interval. A value of p ≤ 0.05 was considered statistically significant.
Results
β-Catenin Accumulation in Monocyte-Derived Dendritic Cells
β-catenin was detectable in immature moDCs (iDCs) by Western blotting, and this concentration increased slightly in moDCs matured for 23 h using LPS (Figure 1A). 6-BIO dose-dependently increased the accumulation of β-catenin when moDCs were treated with 6-BIO for 24 h and with concomitant LPS for the last 23 h prior to cell harvesting according to Western blot analyses (Figure 1A). Indirect fluorescent confocal microscopy revealed β-catenin accumulation in the cell nuclei of moDCs treated with 0.25 to 1 μM 6-BIO (Figure 1B and Supplementary Figure 1).
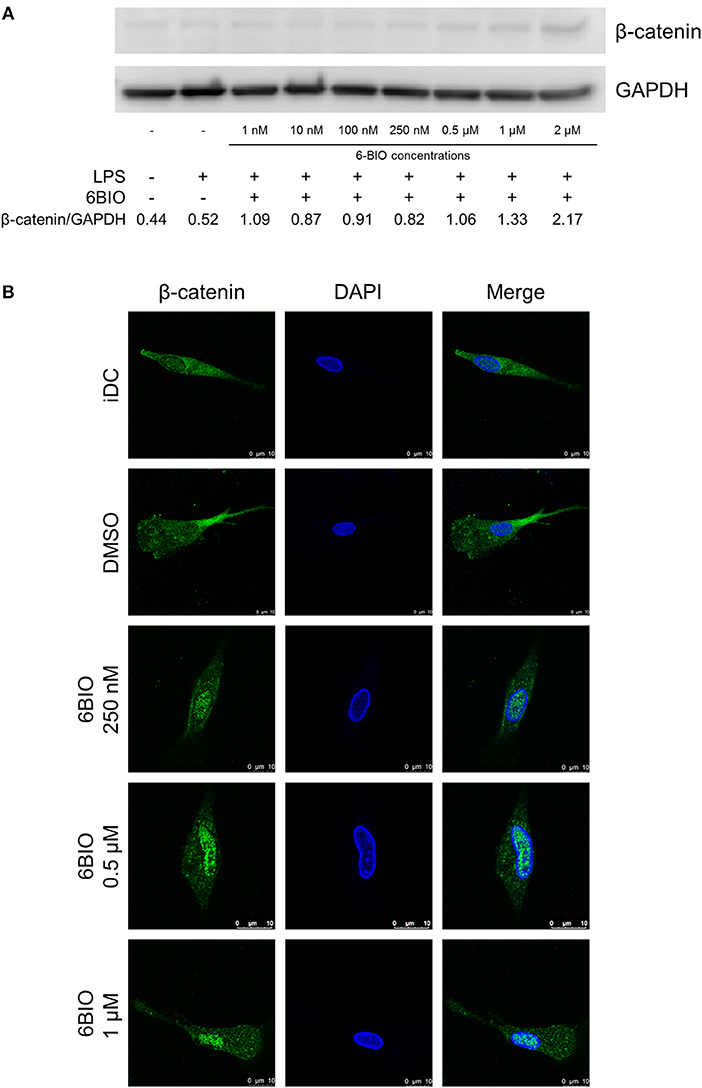
Figure 1. Monocyte-derived dendritic cells (moDCs) obtained from healthy donors were treated with indicated concentrations of 6-bromoindirubin-3′-oxime (6-BIO) or DMSO (vehicle) for 24 h together with 30 ng/ml of lipopolysaccharide (LPS) for the last 23 h or left untreated as iDC. (A) β-catenin protein levels were measured by Western blot in whole-cell lysates. Representative Western blot of three independent experiments is shown. (B) Fluorescein (FITC) indirect immunofluorescent detection of β-catenin proteins according to confocal microscopy analyses.
6-BIO Dose-Dependently Promoted Anti-Inflammatory Features of moDCs
In order to further investigate possible immune-relevant consequences of 6-BIO-induced β-catenin in mature moDCs, we quantified IL-12p70 and IL-10 of cell culture supernatants using ELISA. As exemplified in Figure 2A, the IL-12 concentration decreased significantly with increasing 6-BIO concentrations following LPS maturation for 23 h. 6-BIO concentrations down to 1 nM decreased IL-12 levels compared to cultures with only vehicle and with pronounced dose-dependent IL-12 decrease at 10, 100, and 250 nM. On the contrary, IL-10 secretion increased dose dependently and significantly with increasing 6-BIO in LPS-matured moDCs (Figure 2B). Absolute levels of IL-10 and IL-12 secretion varied considerably in moDCs of buffy coats donated by different healthy persons, although the above trends were in common, for which reason fold change was used for the Y-axis of Figure 2. Quantitative levels in picogram/milliliter are shown in Supplementary Figure 2.
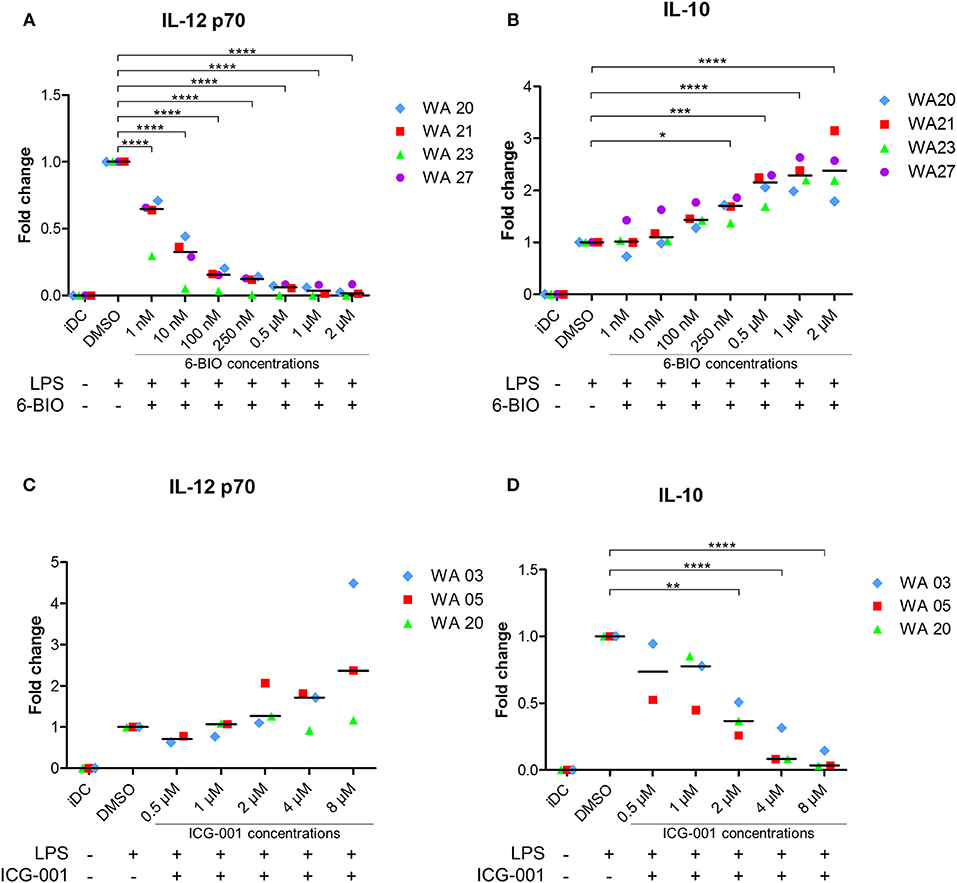
Figure 2. moDCs obtained from healthy donors were treated with indicated concentrations of 6-BIO or ICG-001 or DMSO for 24 h with 30 ng/ml of LPS for the last 23 h or left untreated with those compounds as iDC. (A–D) The fold changes compared to DMSO controls of IL-12p70 and IL-10 in supernatants were measured by enzyme-linked immunosorbent assay (ELISA). Each symbol represents a different donor, and lines represent the median. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 using one-way ANOVA with Dunnett's multiple comparisons test and 95% confidence interval.
ICG-001 Increased IL-12 and Decreased IL-10 Secretion of Mature moDCs
In order to examine the possibility that β-catenin signaling was activated in LPS-matured moDCs even without the use of 6-BIO, cells were treated with the commercially available β-catenin inhibitor ICG-001 for 24 h and with LPS added for the last 23 h before harvest of supernatants. ICG-001 between 1 and 8 μM dose-dependently increased the secretion of IL-12p70 (Figure 2C). In the same supernatants, a dose-dependent decrease in IL-10 was found with significantly increasing effect from 1 to 8 μM ICG-001 (Figure 2D).
6-BIO and ICG-001 Competed Against Each Other Regarding Pro- and Anti-Inflammatory Features
To examine any direct competition between 6-BIO and ICG-001, ELISA was used to quantify IL-12 and IL-10 secretion of LPS-matured DCs (Figure 3). IL-12 induced by either LPS alone or by LPS plus 8 μM ICG-001 was dose-dependently competed by 6-BIO (Figure 3A). Correspondingly, IL-10 induced by either LPS alone or by LPS plus 6-BIO was efficiently competed by 8 μM ICG-001 (Figure 3B). Western blots showed that ICG-001 did not significantly affect the 6-BIO-induced accumulation of β-catenin (Figure 3C). Quantification of mRNA levels using Agilent microarray and RNA-seq profiling showed that β-catenin mRNA levels remained relatively unaffected by ICG-001 treatment and was reduced by possible negative feedback following 6-BIO treatment (Supplementary Figure 3).
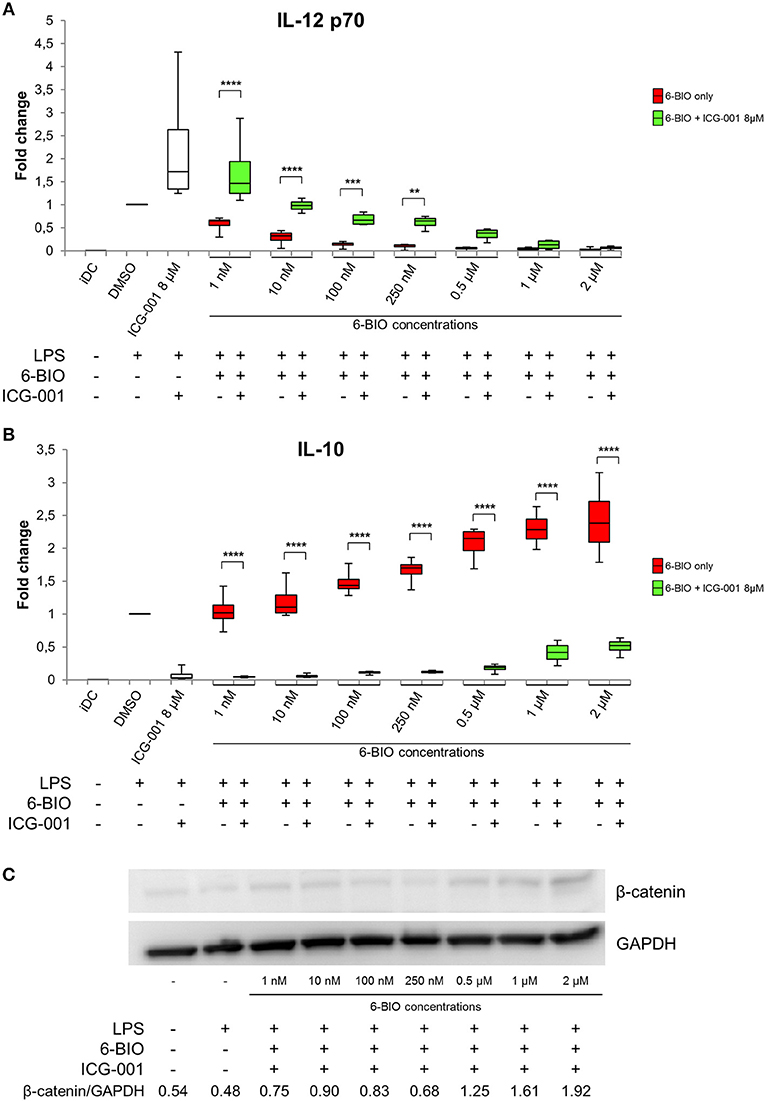
Figure 3. moDCs obtained from healthy donors were treated with 8 μM ICG-001 and/or indicated concentrations of 6-BIO or DMSO (vehicle) for 24 h with 30 ng/ml of LPS for the last 23 h or left untreated as iDC. (A,B) Each vertical box and whisker plot shows the fold change compared to untreated DMSO controls of IL-12p70 and IL-10, respectively, measured by ELISA. The lines represent the median, edges of the box represent 25th and 75th percentiles, and whiskers display the smallest and highest value, n = 5. (C) β-catenin protein levels were measured by Western blot in whole-cell lysates. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 using two-way ANOVA with Tukey's multiple comparisons test and 95% confidence interval.
Effect of 6-BIO and ICG-001 on Maturation and Activation Markers of DCs
Flow cytometry was used to examine how relevant DC surface markers were affected by β-catenin stimulation or inhibition. A representative gating strategy is shown in Supplementary Figure 4. Retained monocyte marker CD14, along with decreased CD1a, suggests deviated maturation of DCs (29). ICG-001 treatment decreased the percentage of LPS-matured DCs that expressed CD14. 6-BIO (100 and 250 nM) increased the CD14 expression, and this was counteracted by the addition of 8 μM ICG-001 (Figure 4). CD1a expression was not affected by either treatment. The maturation markers HLA-DR and CD83 showed, as expected, a clear upregulation following LPS treatment for 23 h. HLA-DR was not strongly affected by either ICG-001 or 6-BIO. 6-BIO was moderately inhibitory and ICG-001 moderately stimulatory to CD83 expression at the concentrations tested (Figure 4).
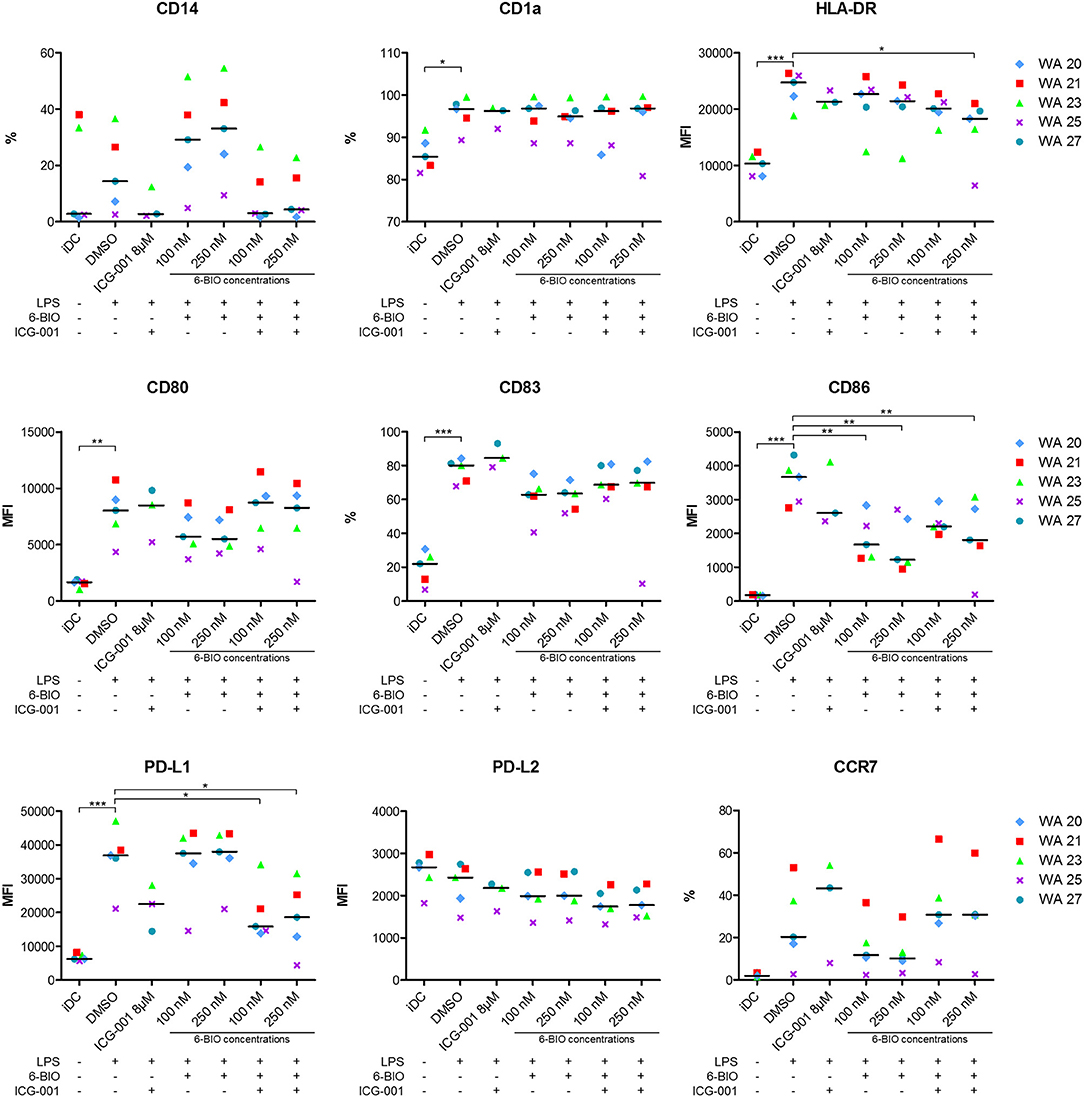
Figure 4. moDCs obtained from healthy donors were treated with 8 μM ICG-001 and/or indicated concentrations of 6-BIO or DMSO for 24 h with 30 ng/ml of LPS for the last 23 h or left untreated as iDC. Flow cytometry analyses of cell surface markers were performed. Each symbol represents a different donor and lines represent the median. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, using one-way ANOVA with Dunnett's multiple comparisons test and 95% confidence interval. %, percentage of positive cells; MFI, median fluorescence intensity.
The activation markers CD80 and CD86 increased strongly as expected when iDCs were stimulated for 23 h with LPS. 6-BIO clearly reduced expression of LPS-induced CD80 and CD86, while the effect of ICG-001 was less clear (Figure 4). The inhibitory marker PD-L1 increased strongly following LPS maturation of iDC for 23 h with or without 6-BIO. ICG-001 (8 μM) tended to decrease the LPS-induced PD-L1 with or without 100 or 250 nM 6-BIO. The PD-L2 marker was not increased by LPS, but was moderately decreased by 8 μM ICG-001 (Figure 4). The DC migration marker CCR7 was upregulated during LPS maturation, as expected, and was clearly reduced by 6-BIO treatment. ICG-001 increased CCR7 expression and competed the 6-BIO-associated downregulation of CCR7.
Effect of 6-BIO and ICG-001 on Gene Expression Patterns
In order to obtain a broader overview of potential pro- and anti-inflammatory features of LPS-matured moDCs, mRNA transcription was profiled using both Agilent 44k microarrays and Illumina RNA-seq. Figure 5 shows a selection of 26 genes known to be relevant in pro- and anti-inflammatory regulation. The performance of the model with gene expression analyses was exemplified by the known β-catenin target gene LRP5 (30). Both Agilent 44k microarrays and Illumina RNA-seq showed a significant, but low level, iDC expression of LRP5 mRNA, with almost 48-fold induction of LRP5 in LPS-matured moDCs treated with 6-BIO. In contrast, LRP5 was reduced in ICG-001-treated LPS-matured moDCs. The classical β-catenin target gene AXIN2 was relatively weakly expressed as mRNA, but with significant induction with concomitant 6-BIO in LPS-matured moDCs (Figures 5A,B). CTNNB1 (β-catenin) mRNA was relatively abundant and was reduced by 6-BIO and little affected by ICG-001 in LPS-matured moDCs.
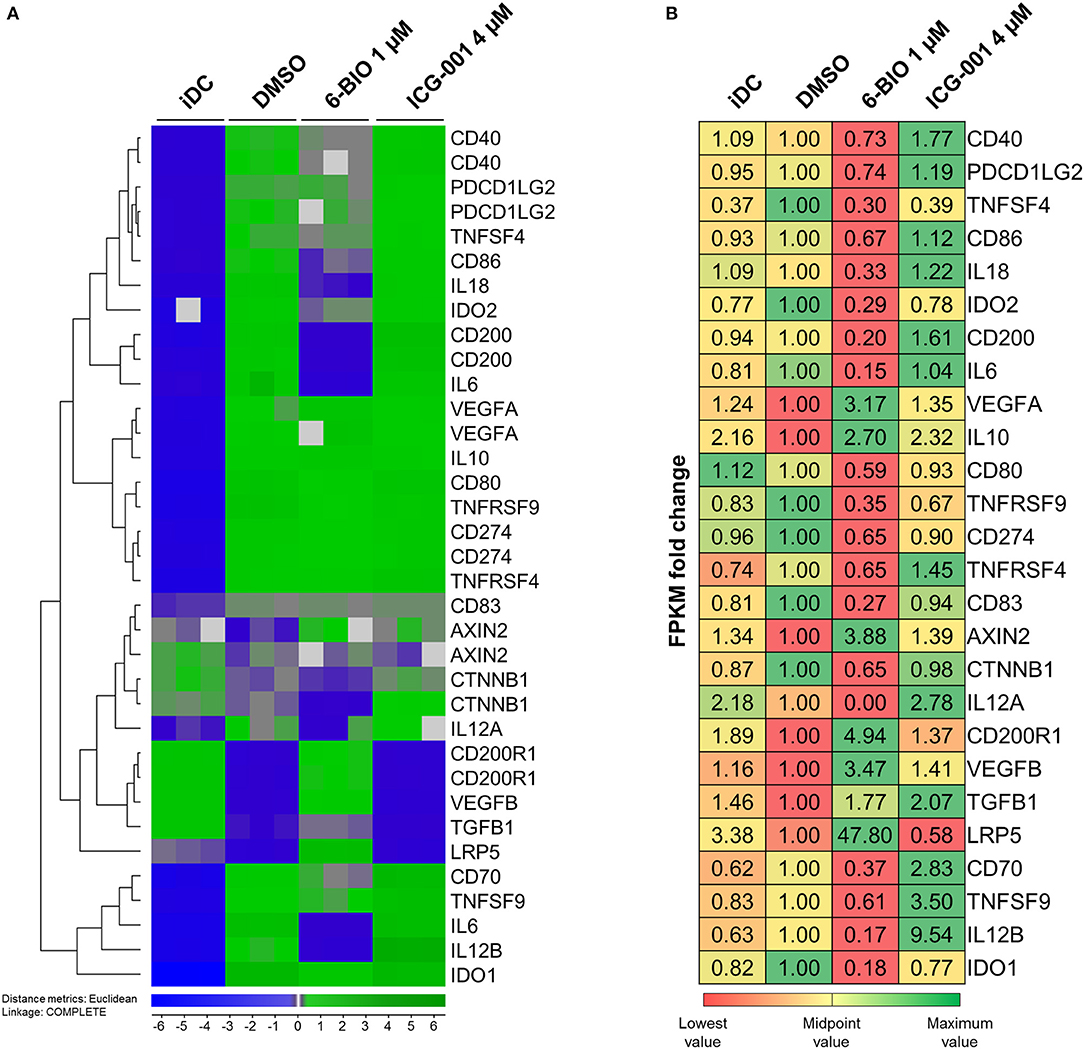
Figure 5. moDCs obtained from healthy donor and treated with DMSO, 1 μM 6-BIO or 8 μM ICG-001 for 24 h with 30 ng/ml of LPS for the last 23 h, or left untreated as iDC. (A) Total RNA was subjected to whole-genome microarray analysis. Heat map of selected gene expression data based on the supervised hierarchical cluster analysis (J-Express™ software) of different treatments. (B) Total RNA (500 ng) was used to generate RNA-seq libraries. Values of the expression of selected genes were based on fold change of normalized fragments per kilobase of transcript per million mapped fragments (FPKM) for each gene of each sample. Fold change was compared to untreated DMSO (vehicle) controls.
Transcription Levels of Pro- and Anti-Inflammatory Interleukins
According to Agilent 44k microarray data (Figure 5A) pro-inflammatory interleukins, IL-6, IL-12B, and IL-18 were strongly induced in LPS-matured moDCs compared to their immature origins. RNA-seq data validated the increase in IL-6 and IL-12B (Figure 5B). Parallel treatment with 1 μM 6-BIO clearly reduced mRNA levels of all of IL-6, IL-12B, IL18, and IL-12A according to both microarray and RNA-seq analyses. ICG-001 (4 μM) induced the opposite effect and a clear increase in these pro-inflammatory mRNA markers in LPS-matured moDCs according to both data sets. The anti-inflammatory IL-10 mRNA was increased in LPS-matured moDCs compared to iDCs according to microarray data, but this was not validated by RNA-seq data. Concomitant 6-BIO-treatment, however, was associated with increased IL-10 mRNA according to both data sets with less clear effect of concomitant ICG-001 treatment.
Transcription Levels of Activating and Inhibitory Membrane Markers
The DC activation markers CD40, CD80, and CD86 were clearly increased in LPS-matured moDCs according to microarrays (Figure 5A). These results were validated by RNA-seq data for CD86, but not for CD40 and CD80. Concomitant 6-BIO and LPS treatment showed consistently lower levels of all these cell surface activation markers according to microarrays in comparison with only LPS and, in particular, with concomitant LPS and ICG-001 treatment. RNA-seq data validated the 6-BIO results for CD40, CD80, and CD86 and showed clearly higher expression levels when ICG-001 was used instead of 6-BIO in LPS-matured moDCs (Figure 5B). DC membrane inhibitory markers PDCD1LG2 (PD-L1) and CD274 (PD-L2) mRNAs were induced in LPS-matured moDCs according to both data sets, while concomitant 6-BIO treatment reduced transcription and ICG-001 increased transcription according to RNA-seq data. TNFS4 (OX40 ligand) is typically expressed on DCs and its receptor TNFRSF4 (OX40) typically on T lymphocytes (31). According to microarray data, TNFRSF4 increased substantially in LPS-matured DCs and was reduced by concomitant 6-BIO and increased by ICG-001 according to both microarrays and RNA-seq (Figures 5A,B). TNFSF9 (4-1BBL; CD137L) expressed on DCs activate lymphocytes via binding to TNFRSF9 (CD137) (32). Microarray data showed strongly induced expression of both TNFSF9 and TNFRSF9 mRNAs in LPS-matured moDCs. Concomitant 6-BIO treatment was associated with lower, and ICG-001 treatment with higher, TNFSF9 expression (Figures 5A,B). CD70 is a cell-membrane-bound TNF superfamily (TNFSR) member that activates T lymphocytes via TNFRSF member CD27 (31). According to microarray data, CD70 mRNA was strongly induced by LPS maturation of moDCs, and this was reduced by 6-BIO and increased by ICG-001 (Figures 5A,B). CD200 is a surface glycoprotein that can induce IDO expression following engagement of pDC-expressed CD200R1. According to microarray data, CD200 mRNA was strongly induced in LPS-matured moDCs. According to both microarray and RNA-seq data, CD200 expression is further increased by concomitant ICG-001, but reduced by concomitant 6-BIO (Figures 5A,B). CD200R1 was clearly reduced in LPS-matured moDCs, but was increased by concomitant 6-BIO treatment (Figures 5A,B).
Transcription Levels of Inhibitory Enzymes and Secreted Proteins
Indoleamine 2,3-dioxygenase (IDO-1) activity, via enzymatic catalysis of tryptophan metabolites, converts mature DCs into tolerogenic antigen-presenting cells that suppress T effector cells and promote T regulatory cells, thereby promoting tolerance (33). IDO1 was found strongly upregulated in LPS-stimulated moDCs. Concomitant 6-BIO reduced IDO1 (Figures 5A,B). TGFB is a secreted immunomodulatory molecule of DCs (10, 31). According to both microarray and RNA-seq data, TGFB1 is reduced in LPS-stimulated moDCs compared to iDCs. Concomitant 6-BIO treatment stimulated TGFB1 expression (Figures 5A,B). VEGF expression and secretion are associated both with aberrant DC maturation and anti-inflammatory DCs (34), and both VEGFA and VEGFB increased strongly in LPS- plus 6-BIO-treated moDCs (Figures 5A,B).
Functional Effects of 6-BIO and ICG-001 in the Allogeneic Mixed Leukocyte Reaction (MLR)
Allogeneic MLRs were performed to analyze T-cell stimulatory capacity of the generated DC populations. The cells were co-cultured with monocyte-depleted allogeneic CFDA-SE-stained PBMCs, and proliferation was determined by the reduction in CFSE intensity. All DC populations showed T-cell stimulatory capacity with LPS-matured moDCs inducing more proliferation than iDC (Figure 6A). The addition of 6-BIO did not, however, show any significant variation in T-cell proliferation compared to DMSO control, although a slight decrease was observed at 0.5 and 2 μM 6-BIO. Neither did up to 8 μM ICG-001 significantly affect the recorded T-cell proliferation (result not shown). In order to examine further the apparent minimal effect of β-catenin modulation of the MLR assay, co-culture supernatants were investigated for cytokines using the Luminex microbead assay (Figure 6B). There was a trend toward 6-BIO dose-dependent reduction in pro-inflammatory cytokines IFN-γ, TNF-α, and IL-6. Among those cytokines, only IL-6 level reached statistical significance. Soluble IL-2R showed a statistically significant and dose-dependent decrease with increasing 6-BIO. It was additionally noted that the 6-BIO dose-dependent effects on IL-10 and IL-12 that we found in pure DC cultures seemed to be abrogated in co-cultures with allogeneic lymphocytes.
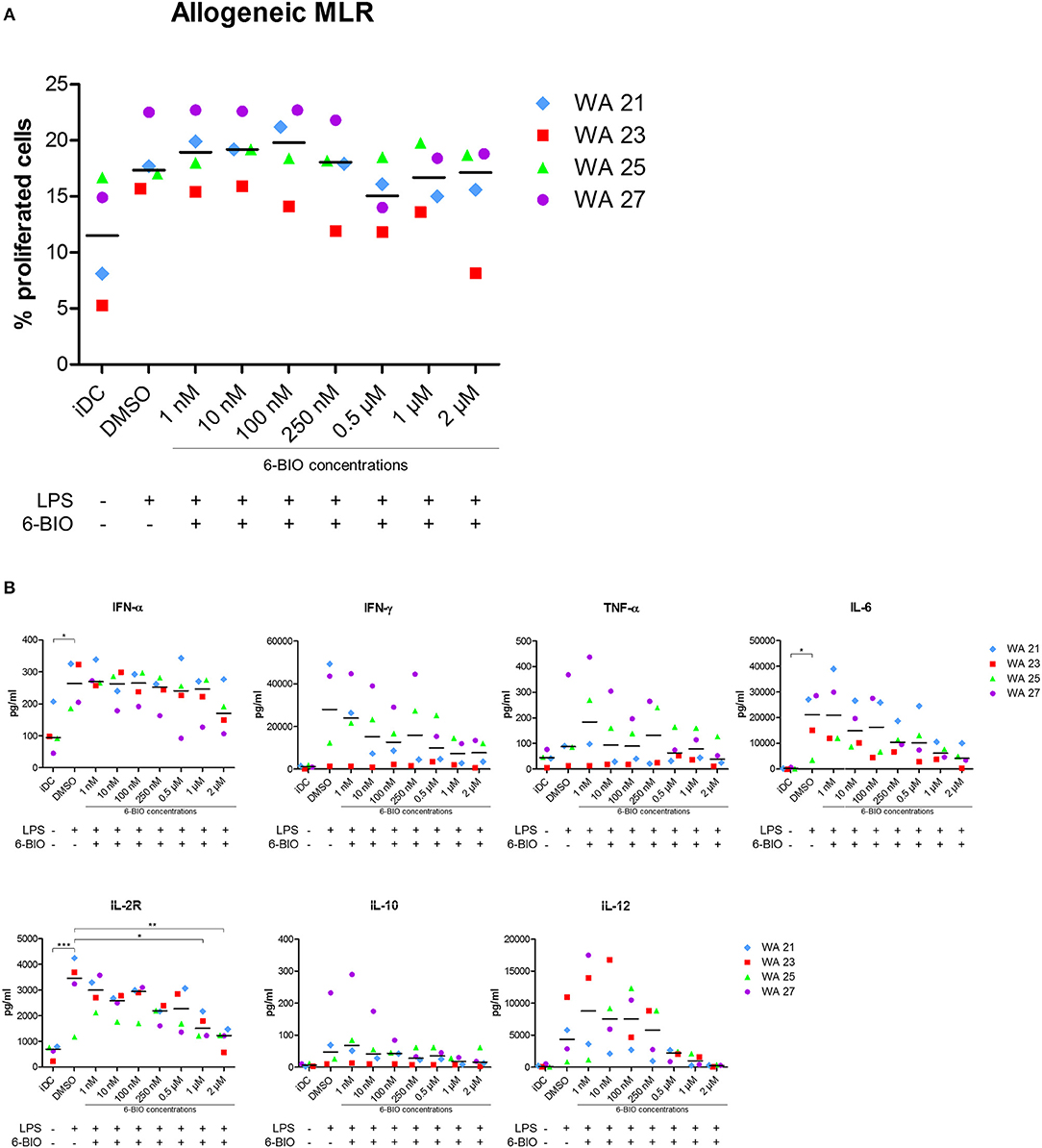
Figure 6. Mixed leukocyte reaction (MLR) between moDCs and allogeneic peripheral blood mononuclear cells (PBMCs) obtained from healthy donors. moDCs were treated with indicated concentrations of 6-BIO or DMSO for 24 h with 30 ng/ml of LPS treatment for 23 h, or left untreated as iDC. Allogeneic PBMCs were pre-labeled with CFSE and co-cultured with treated moDCs for 5 days. (A) Percentage proliferated cells was measured by reduction in CFSE intensity. (B) The amount of secreted cytokines was analyzed using 25-Plex Panel, and the concentrations are given in pg/ml. Each symbol represents a different donor, and lines represent the median. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, using one-way ANOVA with Dunnett's multiple comparisons test and 95% confidence interval.
Discussion
Improved DCs are much needed for next-generation cancer immunotherapy (2, 7, 8, 35). One attractive possibility is that β-catenin signaling can be exploited to generate more robust and potent therapeutic DCs. The transcription factor β-catenin is an important immune regulator that can affect pro-inflammatory and anti-inflammatory/tolerogenic features of mouse and human DCs of different subtypes (11, 12, 36–40), but the therapeutic potential of β-catenin inhibition in moDCs needs further clarification (41–47).
In the present work, we established a model system of moDCs derived from buffy coats of healthy blood donors. Monocytes were induced to become immature DCs (iDCs) using GM-CSF and IL-4 for 4 days. Thereafter, small molecular compounds were added to iDCs for 24 h with concomitant LPS for the last 23 hours, followed by assays of secreted IL-10 and IL-12p70 and surface markers of maturation, activation, and inhibition.
In order to investigate the possibility that a basal β-catenin activation was present in LPS-matured moDCs (48), even in the absence of 6-BIO, we tested our model with the β-catenin inhibitor ICG-001, previously found to inhibit β-catenin-stimulated transcription in both cancer cells (49) and dendritic cells (42). A pronounced ICG-001 dose-dependent increase in IL-12p70 and a corresponding dose-dependent decrease in IL-10 secretion was observed when iDCs were treated for 24 h with ICG-001. This suggested that β-catenin is activated in mature moDCs to promote anti-inflammatory features and, furthermore, is accessible to β-catenin inhibition, even in the absence of Wnt ligands or other specific activators of Wnt or β-catenin signaling added to the culture medium.
Addition of the small molecular compound 6-BIO indicated, however, that the basal β-catenin activity of LPS-matured moDCs can be augmented by external stimulation. Western blot analyses of moDC whole-cell lysates detected β-catenin both in iDCs and LPS-matured moDCs with dose-dependent increase in β-catenin accumulation detectable from low 6-BIO concentrations. Confocal microscopy revealed obvious nuclear accumulation of β-catenin at 250 nM and higher concentrations of 6-BIO. This in situ localization method was preferred over nuclear and cytoplasmic fractionation, consistent with our previous results that fractionation is associated with loss of labile nuclear proteins due to the drop of colloid osmotic pressure during fractionation (50).
The small molecule ICG-001 binds CREB-binding protein (CBP) to disrupt its interaction with β-catenin and inhibits CBP function as a co-activator of Wnt/β-catenin-mediated transcription (17). This mechanism of β-catenin inhibition is consistent with our present findings. According to our Western blot assays, ICG-001 did not affect β-catenin at the protein synthesis/stability level. According to microarray and RNA-seq data, β-catenin (CTNNB1) mRNA levels were similar in ICG-treated and non-treated moDCs. ICG-001 was consequently used to examine further the association between β-catenin inhibition and anti-inflammatory features of mature moDCs. Competition assays between 6-BIO and ICG-001 showed that these compounds had inverse effects on IL-10 and IL-12p70 secretion and on dendritic cell markers of activation, inhibition, and migration. 6-BIO dose-dependently promoted anti-inflammatory patterns of the examined cytokines and several surface markers. ICG-001 exhibited the opposite and the competing effect on cytokine and several surface markers. The pronounced upregulation of the β-catenin target gene LRP5 in 6-BIO-treated and downregulation in ICG-001-treated LPS-matured moDCs additionally support the existence of a basal and dynamic β-catenin activation status.
IL-10 is considered an anti-inflammatory cytokine that is induced by β-catenin signaling (11) and by non-canonical Wnt5a signaling in mouse DCs (39). Stimulation of toll-like-receptors (TLR2, TLR4, TLR5, TLR7, and TLR9) has been shown to induce IL-10 production of DCs [reviewed in (14)]. The TLR4 stimulator LPS induced dual production of IL-10 and IL-12 in human moDCs, but with significant donor-to-donor variation (13, 51), consistent with our present findings. In the LPS-matured moDCs, the level of β-catenin was dose-dependently increased by 6-BIO. It may offer experimental advantages to activate β-catenin downstream of the cytoplasmic destruction complex because several upstream and parallel pathways, such as WNTs, tankyrases (52, 53), and TLR2 stimulation via PI3 kinase and ERK in moDCs (54, 55) may activate nuclear β-catenin signaling. In parallel with increasing β-catenin levels, 6-BIO dose-dependently increased IL-10 and dose-dependently decreased IL-12p70. These experiments, therefore, reveal the potential of β-catenin signaling to modulate important pro- and anti-inflammatory cytokine production of moDCs. The IL-10 promoter has been shown to contain several response elements that can bind β-catenin to activate IL-10 transcription (56). Both microarray and RNA-seq data showed increased IL-10 mRNA in 6-BIO-treated moDCs. The relationship between β-catenin signaling and IL-12 stimulation is additionally complex due to the ability of IL-10 to decrease IL-12 production in DCs (13).
In mice, DC-specific deletion of the Wnt co-receptors LRP5/6 or β-catenin led to an increased expression of IL-6, TNF-α, IL-1β, IL-12p40, and IL-12p70 with diminished production of IL-10 and TGF-β (57). In another study on mouse splenic DC precursors, CD11c-specific constitutive β-catenin activation upregulated Irf8 through targeting of the Irf8 promoter, β-catenin-stabilized CD8a + DCs secreted elevated IL-12 upon in vitro microbial stimulation, and pharmacological β-catenin inhibition using ICG-001 blocked this response in wild-type cells (42). Also, knock-down of the non-canonical Wnt5a in human moDCs, presumed not to signal via β-catenin, compromised IL-12 secretion (58).
In order to obtain an additional impression of potential pro- and anti-inflammatory features of LPS-matured moDCs, whole-genome mRNA analyses of moDCs was done, using both Agilent microarray and Illumina RNA sequence quantifications. Recently, it was published that β-catenin directly stimulates the IDO1 transcription and additionally the peroxisome proliferator-activated receptor-γ (PPAR-γ), thus, causing the metabolic shift required for protoporphyrin X synthesis, the heme prosthetic group required for full IDO1 enzymatic activity in DCs (12). According to our mRNA expression data, IDO1 was strongly induced and abundantly expressed in LPS-matured moDCs, while PPARG was relatively abundantly expressed in both immature and LPS-matured DCs (ArrayExpress E-MTAB-8330). Surprisingly, 6-BIO dose-dependently reduced IDO1 mRNA levels in our LPS-matured moDCs, although significant IDO1 mRNA was still expressed even at high 6-BIO concentrations. Interestingly, CD200 binding to CD200R1 on murine pDCs has been reported among stimulators of IDO1 expression (59). Expressions of CD200 and CD200R1 were strongly affected during LPS maturation and by concomitant 6-BIO and ICG-001 treatment. These findings have to be followed up separately.
It has been shown in both murine and human models that Wnt5a from melanomas affects the local DCs to express indoleamine 2,3-dioxygenase-1 (IDO1) that stimulates development of T regulatory cells (TRegs) through kynurenine (60). Wnt5a has been considered to be a ligand of the non-canonical Wnt pathway (β-catenin independent), but appears to be able to context-dependent stimulation of β-catenin signaling, in some cases stronger than the “canonical” Wnt3a stimulation (12, 60).
TGF-β induces T regulatory cells and thereby promotes tolerance when secreted from antigen-presenting cells [(10, 31, 48) and references therein]. It has been reported that TGF-β antagonizes β-catenin in DCs, thereby selectively suppressing signaling associated with tolerogenic DC activation while having no impact on LPS-induced, β-catenin-independent immunogenic activation (61). According to our microarray and RNA-seq data, TGFB1 was clearly reduced during LPS-mediated maturation of moDCs, but this decrease was counteracted by both concomitant 6-BIO and ICG-001 treatment. This could represent indirect effects of β-catenin, also because TGFB1 is not a known β-catenin target gene.
VEGF has been shown to be a direct β-catenin target gene in different cell types (62). The strong 6-BIO-enhanced expression of both VEGFA and VEGFB would suggest this to be the case in LPS-matured moDCs. VEGF has been shown to be immunosuppressive in different ways: it can inhibit the function of T cells, increase the recruitment of T regulatory cells and myeloid-derived suppressor cells (MDSCs), and hinder the differentiation and activation of DCs (34).
Additional transcriptional determinants of tolerogenic and immunogenic states during dendritic cell maturation have been published (63). All our genome-wide Agilent microarray and Illumina RNA-seq data have been made publicly available and can be further explored regarding β-catenin targets and pro- and anti-inflammatory transcription of LPS-matured moDCs.
Mixed leukocyte reaction (MLR) was employed for the assessment of T-cell stimulatory capacity of the generated cell populations and showed the ability of the LPS-stimulated moDCs to induce allogeneic T-cell proliferation. The minor effects of either 6-BIO or ICG-001 in the MLR assay could reflect that the outcome of conflicting pro- and anti-inflammatory cues could be complex. It is possible that negative feedback effects due to moDC and allogeneic cross-talk could result in the abrogation of the pronounced dose-dependent effects on both IL-10 and IL-12 secretion by either 6-BIO or ICG-001 in pure moDC. Expanded cytokine profiling of MLR culture supernatants showed 6-BIO dose-dependent decreases of several relevant cytokines. Presently, this is a reminder of increased complexity once different immune cells are brought into interaction. Future work will address such interactions in MLR assays and in DC and patient-derived cell co-cultures with expanded assays including parallel multi-variable mass cytometric analyses of supernatants and cells.
Much understanding is lacking regarding β-catenin signaling in moDCs, although a critical role of β-catenin signaling in DC function and differentiation of pro- and anti-inflammatory features in vivo is already established [reviewed in (37, 46)]. In both freshly isolated and Flt3-stimulated CD11c+ DCs from mouse lymph nodes, the main conclusion was that Wnts upregulate immune suppressive cytokines (IL-10, VEGF, TGF-β) without inhibiting LPS-induced maturation and activation, thus allowing development of a mature tolerogenic phenotype (39). Minimal effects were detected on the MHCII maturation marker or CD80 or CD86 activation markers or the migration marker CCR7 (39). IL-12p70 secretion was additionally found to be little affected by either canonical (Wnt3a) or non-canonical (Wnt5a) signaling in that study (39). In another study, however, deletion of β-catenin in a mouse model was found to increase expression of DC co-stimulatory markers (CD40, CD80, CD86) and to decrease the inhibitory markers PD-L1 and PD-L2 (64). Different murine tumor models have documented the ability of Wnt ligands to stimulate DCs to produce tolerogenic factors, such as IL-10, Raldh, and Ido-1 [reviewed in (11)]. In one study of human cancer, melanoma-intrinsic β-catenin signaling was found to inhibit DC migration and lead to immune evasion (65). The important mechanisms involved have been reviewed (38, 40, 66, 67).
Conclusion
LPS-matured moDCs co-developed pro- and anti-inflammatory surface and secretory markers with considerable quantitative person-to-person variation. A basal β-catenin activation was present in LPS-matured moDCs and could be boosted dose dependently by the β-catenin activator 6-BIO and counter-acted dose-dependently by the β-catenin transcription complex inhibitor ICG-001 with inverse effects on IL-10 and IL-12 secretion. These observations should be taken into consideration for the production of more potent and robust therapeutic DCs.
Data Availability Statement
DNA microarray data have been deposited into the ArrayExpress database under accession number E-MTAB-8330.
Author Contributions
WA, RB, and AØ did the experiments. WA prepared the figures. All authors contributed to experimental design, evaluation of results, manuscript revisions, and approved the submitted version.
Funding
The Research Council of Norway (Norges Forskningsråd) has funded positions and running costs. The Norwegian Cancer Society (Kreftforeningen) has funded positions and running costs. This work received funding from Helse Vest (grant numbers 912062, 911626, 911747, 911582, 911778, 912226, and 980058), the Helse Vest Strategic grants of Personalized Therapy (grant number 303484) and Bergen Stem Cell Consortium (grant number 502027), Centre for Cancer Biomarkers, CCBIO, a Centre of Excellence in Cancer Biomarkers authorized and funded by the Research Council of Norway, the Research Council of Norway NFR BEHANDLING program (project number 287829); the Norwegian Cancer Society (project number 204947) and Bergen Research Foundation/Trond Mohn Stiftelse. All flow cytometry experiments were performed at the Core Facility for Flow Cytometry, Department of Clinical Science, University of Bergen.
Conflict of Interest
AØ and K-HK own stocks in the Company Alden Cancer Therapy II AS that has sponsored and will sponsor dendritic cell-based cancer immunotherapy, including the trial registered at ClinicalTrials.gov Identifier: NCT02423928. AØ and K-HK additionally are co-inventors of the United States Patent Application No. 15/771496 (US-2018-0344715) regarding novel beta-catenin inhibitor compounds for cancer therapy. None of these compounds have been used or addressed in the present manuscript.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Hua My Hoang is acknowledged for expert technical assistance and Kjell Petersen and the Computational Biology Unit, Department of Informatics, University of Bergen, for J-Express and microarray data handling. Illumina RNA-sequencing was done by Qiagen GmbH, Hilden, Germany. We thank Michael Gombert and Charlotte B. Ahler for help in data analyses. We thank blood donors and staff at the Blood bank, Haukeland University Hospital. We greatly acknowledge the contributions to this study by Einar Galtung Døsvig, Espen Galtung Døsvig, Bjarne Rieber, Trond Mohn, Herman Friele, Jan Einar Greve, Kåre Rommetveit, and Thorstein Selvik.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.00438/full#supplementary-material
References
1. Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. (1998) 392:245–52. doi: 10.1038/32588
2. Bol KF, Schreibelt G, Rabold K, Wculek SK, Schwarze JK, Dzionek A, et al. The clinical application of cancer immunotherapy based on naturally circulating dendritic cells. J Immunother Cancer. (2019) 7:109. doi: 10.1186/s40425-019-0580-6
3. Chrisikos TT, Zhou Y, Slone N, Babcock R, Watowich SS, Li HS. Molecular regulation of dendritic cell development and function in homeostasis, inflammation, and cancer. Mol Immunol. (2019) 110:24–39. doi: 10.1016/j.molimm.2018.01.014
4. Collin M, Bigley V. Human dendritic cell subsets: an update. Immunology. (2018) 154:3–20. doi: 10.1111/imm.12888
5. Alcantara-Hernandez M, Leylek R, Wagar LE, Engleman EG, Keler T, Marinkovich MP, et al. High-dimensional phenotypic mapping of human dendritic cells reveals interindividual variation and tissue specialization. Immunity. (2017) 47:1037–50.e6. doi: 10.1016/j.immuni.2017.11.001
6. Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. (2017) 356:eaah4573. doi: 10.1126/science.aah4573
7. Sabado RL, Balan S, Bhardwaj N. Dendritic cell-based immunotherapy. Cell Res. (2017) 27:74–95. doi: 10.1038/cr.2016.157
8. Saxena M, Bhardwaj N. Re-emergence of dendritic cell vaccines for cancer treatment. Trends Cancer. (2018) 4:119–37. doi: 10.1016/j.trecan.2017.12.007
9. Jiang A, Bloom O, Ono S, Cui W, Unternaehrer J, Jiang S, et al. Disruption of E-cadherin-mediated adhesion induces a functionally distinct pathway of dendritic cell maturation. Immunity. (2007) 27:610–24. doi: 10.1016/j.immuni.2007.08.015
10. Manicassamy S, Pulendran B. Dendritic cell control of tolerogenic responses. Immunol Rev. (2011) 241:206–27. doi: 10.1111/j.1600-065X.2011.01015.x
11. Suryawanshi A, Tadagavadi RK, Swafford D, Manicassamy S. Modulation of inflammatory responses by Wnt/beta-catenin signaling in dendritic cells: a novel immunotherapy target for autoimmunity and cancer. Front Immunol. (2016) 7:460. doi: 10.3389/fimmu.2016.00460
12. Zhao F, Xiao C, Evans KS, Theivanthiran T, DeVito N, Holtzhausen A, et al. Paracrine Wnt5a-beta-catenin signaling triggers a metabolic program that drives dendritic cell tolerization. Immunity. (2018) 48:147–60.e7. doi: 10.1016/j.immuni.2017.12.004
13. Zheng H, Ban Y, Wei F, Ma X. Regulation of interleukin-12 production in antigen-presenting cells. Adv Exp Med Biol. (2016) 941:117–38. doi: 10.1007/978-94-024-0921-5_6
14. Rutz S, Ouyang W. Regulation of interleukin-10 expression. Adv Exp Med Biol. (2016) 941:89–116. doi: 10.1007/978-94-024-0921-5_5
15. Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. (2004) 10:55–63. doi: 10.1038/nm979
16. Qu Y, Gharbi N, Yuan X, Olsen JR, Blicher P, Dalhus B, et al. Axitinib blocks Wnt/beta-catenin signaling and directs asymmetric cell division in cancer. Proc Natl Acad Sci USA. (2016) 113:9339–44. doi: 10.1073/pnas.1604520113
17. Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected]. Proc Natl Acad Sci USA. (2004) 101:12682–7. doi: 10.1073/pnas.0404875101
18. Olsen JR, Azeem W, Hellem MR, Marvyin K, Hua Y, Qu Y, et al. Context dependent regulatory patterns of the androgen receptor and androgen receptor target genes. BMC Cancer. (2016) 16:377. doi: 10.1186/s12885-016-2453-4
19. Azeem W, Hellem MR, Olsen JR, Hua Y, Marvyin K, Qu Y, et al. An androgen response element driven reporter assay for the detection of androgen receptor activity in prostate cells. PLoS ONE. (2017) 12:e0177861. doi: 10.1371/journal.pone.0177861
20. Sprater F, Azeem W, Appel S. Activation of peroxisome proliferator-activated receptor gamma leads to upregulation of ESE-3 expression in human monocyte-derived dendritic cells. Scand J Immunol. (2014) 79:20–6. doi: 10.1111/sji.12126
21. Qu Y, Oyan AM, Liu R, Hua Y, Zhang J, Hovland R, et al. Generation of prostate tumor-initiating cells is associated with elevation of reactive oxygen species and IL-6/STAT3 signaling. Cancer Res. (2013) 73:7090–100. doi: 10.1158/0008-5472.CAN-13-1560
22. Dysvik B, Jonassen I. J-Express: exploring gene expression data using Java. Bioinformatics. (2001) 17:369–70. doi: 10.1093/bioinformatics/17.4.369
23. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. (2012) 9:357–9. doi: 10.1038/nmeth.1923
24. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. (2009) 25:1105–11. doi: 10.1093/bioinformatics/btp120
25. Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Nat Biotechnol. (2009) 27:455–7. doi: 10.1038/nbt0509-455
26. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. (2012) 7:562–78. doi: 10.1038/nprot.2012.016
27. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. (2010) 28:511–5. doi: 10.1038/nbt.1621
28. Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. (2011) 12:R22. doi: 10.1186/gb-2011-12-3-r22
29. Wolfle SJ, Strebovsky J, Bartz H, Sahr A, Arnold C, Kaiser C, et al. PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur J Immunol. (2011) 41:413–24. doi: 10.1002/eji.201040979
30. Hong Y, Manoharan I, Suryawanshi A, Shanmugam A, Swafford D, Ahmad S, et al. Deletion of LRP5 and LRP6 in dendritic cells enhances antitumor immunity. Oncoimmunology. (2016) 5:e1115941. doi: 10.1080/2162402X.2015.1115941
31. Bourque J, Hawiger D. Immunomodulatory bonds of the partnership between dendritic cells and T cells. Crit Rev Immunol. (2018) 38:379–401. doi: 10.1615/CritRevImmunol.2018026790
32. Chu DT, Bac ND, Nguyen KH, Tien NLB, Thanh VV, Nga VT, et al. An update on anti-CD137 antibodies in immunotherapies for cancer. Int J Mol Sci. (2019) 20:1822. doi: 10.3390/ijms20081822
33. Mellor AL, Lemos H, Huang L. Indoleamine 2,3-dioxygenase and tolerance: where are we now? Front Immunol. (2017) 8:1360. doi: 10.3389/fimmu.2017.01360
34. Yang J, Yan J, Liu B. Targeting VEGF/VEGFR to modulate antitumor immunity. Front Immunol. (2018) 9:978. doi: 10.3389/fimmu.2018.00978
35. Saxena M, Balan S, Roudko V, Bhardwaj N. Towards superior dendritic-cell vaccines for cancer therapy. Nat Biomed Eng. (2018) 2:341–6. doi: 10.1038/s41551-018-0250-x
36. Fu C, Jiang A. Dendritic cells and CD8 T cell immunity in tumor microenvironment. Front Immunol. (2018) 9:3059. doi: 10.3389/fimmu.2018.03059
37. Galluzzi L, Spranger S, Fuchs E, Lopez-Soto A. WNT signaling in cancer immunosurveillance. Trends Cell Biol. (2019) 29:44–65. doi: 10.1016/j.tcb.2018.08.005
38. Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. WNT/β-catenin pathway activation correlates with immune exclusion across human cancers. Clin Cancer Res. (2019). 25:3074–83. doi: 10.1158/1078-0432.CCR-18-1942
39. Oderup C, LaJevic M, Butcher EC. Canonical and noncanonical Wnt proteins program dendritic cell responses for tolerance. J Immunol. (2013) 190:6126–34. doi: 10.4049/jimmunol.1203002
40. Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer. (2018) 18:139–47. doi: 10.1038/nrc.2017.117
41. Alessandrini A, De Haseth S, Fray M, Miyajima M, Colvin RB, Williams WW, et al. Dendritic cell maturation occurs through the inhibition of GSK-3beta. Cell Immunol. (2011) 270:114–25. doi: 10.1016/j.cellimm.2011.04.007
42. Cohen SB, Smith NL, McDougal C, Pepper M, Shah S, Yap GS, et al. Beta-catenin signaling drives differentiation and proinflammatory function of IRF8-dependent dendritic cells. J Immunol. (2015) 194:210–22. doi: 10.4049/jimmunol.1402453
43. Feng M, Jin JQ, Xia L, Xiao T, Mei S, Wang X, et al. Pharmacological inhibition of beta-catenin/BCL9 interaction overcomes resistance to immune checkpoint blockades by modulating Treg cells. Sci Adv. (2019) 5:eaau5240. doi: 10.1126/sciadv.aau5240
44. Fu C, Liang X, Cui W, Ober-Blobaum JL, Vazzana J, Shrikant PA, et al. β-Catenin in dendritic cells exerts opposite functions in cross-priming and maintenance of CD8+ T cells through regulation of IL-10. Proc Natl Acad Sci USA. (2015) 112:2823–8. doi: 10.1073/pnas.1414167112
45. Kafer R, Usanova S, Montermann E, Loquai C, Reske-Kunz AB, Bros M. Inhibitors of beta-catenin affect the immuno-phenotype and functions of dendritic cells in an inhibitor-specific manner. Int Immunopharmacol. (2016) 32:118–24. doi: 10.1016/j.intimp.2016.01.018
46. Wang B, Tian T, Kalland KH, Ke X, Qu Y. Targeting Wnt/β-catenin signaling for cancer immunotherapy. Trends Pharmacol Sci. (2018) 39:648–58. doi: 10.1016/j.tips.2018.03.008
47. Zizzari IG, Napoletano C, Botticelli A, Caponnetto S, Calabro F, Gelibter A, et al. TK inhibitor pazopanib primes DCs by downregulation of the beta-catenin pathway. Cancer Immunol Res. (2018) 6:711–22. doi: 10.1158/2326-6066.CIR-17-0594
48. Manicassamy S, Reizis B, Ravindran R, Nakaya H, Salazar-Gonzalez RM, Wang YC. Activation of beta-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science. (2010) 329:849–53. doi: 10.1126/science.1188510
49. Ma H, Nguyen C, Lee KS, Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. (2005) 24:3619–31. doi: 10.1038/sj.onc.1208433
50. Kalland KH, Szilvay AM, Brokstad KA, Saetrevik W, Haukenes G. The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol Cell Biol. (1994) 14:7436–44. doi: 10.1128/MCB.14.11.7436
51. Lovgren T, Sarhan D, Truxova I, Choudhary B, Maas R, Melief J, et al. Enhanced stimulation of human tumor-specific T cells by dendritic cells matured in the presence of interferon-gamma and multiple toll-like receptor agonists. Cancer Immunol Immunother. (2017) 66:1333–44. doi: 10.1007/s00262-017-2029-4
52. Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. (2017) 169:985–99. doi: 10.1016/j.cell.2017.05.016
53. Wiese KE, Nusse R, van Amerongen R. Wnt signalling: conquering complexity. Development. (2018) 145:dev165902. doi: 10.1242/dev.165902
54. Agrawal S, Agrawal A, Doughty B, Gerwitz A, Blenis J, Van Dyke T, et al. Cutting edge: different Toll-like receptor agonists instruct dendritic cells to induce distinct Th responses via differential modulation of extracellular signal-regulated kinase-mitogen-activated protein kinase and c-Fos. J Immunol. (2003) 171:4984–9. doi: 10.4049/jimmunol.171.10.4984
55. Manoharan I, Hong Y, Suryawanshi A, Angus-Hill ML, Sun Z, Mellor AL, et al. TLR2-dependent activation of beta-catenin pathway in dendritic cells induces regulatory responses and attenuates autoimmune inflammation. J Immunol. (2014) 193:4203–13. doi: 10.4049/jimmunol.1400614
56. Yaguchi T, Goto Y, Kido K, Mochimaru H, Sakurai T, Tsukamoto N, et al. Immune suppression and resistance mediated by constitutive activation of Wnt/beta-catenin signaling in human melanoma cells. J Immunol. (2012) 189:2110–7. doi: 10.4049/jimmunol.1102282
57. Suryawanshi A, Manoharan I, Hong Y, Swafford D, Majumdar T, Taketo MM, et al. Canonical wnt signaling in dendritic cells regulates Th1/Th17 responses and suppresses autoimmune neuroinflammation. J Immunol. (2015) 194:3295–304. doi: 10.4049/jimmunol.1402691
58. Valencia J, Martinez VG, Hidalgo L, Hernandez-Lopez C, Canseco NM, Vicente A, et al. Wnt5a signaling increases IL-12 secretion by human dendritic cells and enhances IFN-gamma production by CD4+ T cells. Immunol Lett. (2014) 162(1 Pt A):188–99. doi: 10.1016/j.imlet.2014.08.015
59. Fallarino F, Asselin-Paturel C, Vacca C, Bianchi R, Gizzi S, Fioretti MC, et al. Murine plasmacytoid dendritic cells initiate the immunosuppressive pathway of tryptophan catabolism in response to CD200 receptor engagement. J Immunol. (2004) 173:3748–54. doi: 10.4049/jimmunol.173.6.3748
60. Holtzhausen A, Zhao F, Evans KS, Tsutsui M, Orabona C, Tyler DS, et al. Melanoma-derived Wnt5a promotes local dendritic-cell expression of IDO and immunotolerance: opportunities for pharmacologic enhancement of immunotherapy. Cancer Immunol Res. (2015) 3:1082–95. doi: 10.1158/2326-6066.CIR-14-0167
61. Vander Lugt B, Beck ZT, Fuhlbrigge RC, Hacohen N, Campbell JJ, Boes M. TGF-beta suppresses beta-catenin-dependent tolerogenic activation program in dendritic cells. PLoS ONE. (2011) 6:e20099. doi: 10.1371/journal.pone.0020099
62. Wang Y, Sang A, Zhu M, Zhang G, Guan H, Ji M, et al. Tissue factor induces VEGF expression via activation of the Wnt/beta-catenin signaling pathway in ARPE-19 cells. Mol Vis. (2016) 22:886–97.
63. Vander Lugt B, Riddell J, Khan AA, Hackney JA, Lesch J, DeVoss J, et al. Transcriptional determinants of tolerogenic and immunogenic states during dendritic cell maturation. J Cell Biol. (2017) 216:779–92. doi: 10.1083/jcb.201512012
64. Hong Y, Manoharan I, Suryawanshi A, Majumdar T, Angus-Hill ML, Koni PA, et al. β-catenin promotes regulatory T-cell responses in tumors by inducing vitamin A metabolism in dendritic cells. Cancer Res. (2015) 75:656–65. doi: 10.1158/0008-5472.CAN-14-2377
65. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. (2015) 523:231–5. doi: 10.1038/nature14404
66. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. (2018) 24:541–50. doi: 10.1038/s41591-018-0014-x
Keywords: monocyte-derived dendritic cell, pro-inflammatory, tolerogenic, immunotherapy, beta-catenin
Citation: Azeem W, Bakke RM, Appel S, Øyan AM and Kalland K-H (2020) Dual Pro- and Anti-Inflammatory Features of Monocyte-Derived Dendritic Cells. Front. Immunol. 11:438. doi: 10.3389/fimmu.2020.00438
Received: 28 October 2019; Accepted: 25 February 2020;
Published: 27 March 2020.
Edited by:
Jordi Ochando, Icahn School of Medicine at Mount Sinai, United StatesReviewed by:
Robert Adam Harris, Karolinska Institutet (KI), SwedenEstanislao Nistal-Villan, San Pablo CEU University, Spain
Copyright © 2020 Azeem, Bakke, Appel, Øyan and Kalland. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Waqas Azeem, waqas.azeem@uib.no; Karl-Henning Kalland, karl.kalland@uib.no
 Waqas Azeem
Waqas Azeem Ragnhild Maukon Bakke
Ragnhild Maukon Bakke Silke Appel
Silke Appel Anne Margrete Øyan
Anne Margrete Øyan Karl-Henning Kalland
Karl-Henning Kalland