Abstract
A deep learning-based image analysis could improve diagnostic accuracy and efficiency in pathology work. Recently, we proposed a deep learning-based detection algorithm for C4d immunostaining in renal allografts. The objective of this study is to assess the diagnostic performance of the algorithm by comparing pathologists’ diagnoses and analyzing the associations of the algorithm with clinical data. C4d immunostaining slides of renal allografts were obtained from two different institutions (100 slides from the Asan Medical Center and 86 slides from the Seoul National University Hospital) and scanned using two different slide scanners. Three pathologists and the algorithm independently evaluated each slide according to the Banff 2017 criteria. Subsequently, they jointly reviewed the results for consensus scoring. The result of the algorithm was compared with that of each pathologist and the consensus diagnosis. Clinicopathological associations of the results of the algorithm with allograft survival, histologic evidence of microvascular inflammation, and serologic results for donor-specific antibodies were also analyzed. As a result, the reproducibility between the pathologists was fair to moderate (kappa 0.36–0.54), which is comparable to that between the algorithm and each pathologist (kappa 0.34–0.51). The C4d scores predicted by the algorithm achieved substantial concordance with the consensus diagnosis (kappa = 0.61), and they were significantly associated with remarkable microvascular inflammation (P = 0.001), higher detection rate of donor-specific antibody (P = 0.003), and shorter graft survival (P < 0.001). In conclusion, the deep learning-based C4d detection algorithm showed a diagnostic performance similar to that of the pathologists.
Similar content being viewed by others
Introduction
Antibody-mediated rejection (ABMR) is an important complication following kidney transplantation, contributing to acute and chronic allograft injury in ~10% of kidney transplant recipients [1]. The diagnosis of ABMR requires three factors: histologic evidence of microvascular injury, presence of donor-specific antibody (DSA) in the serologic test, and C4d deposition in the peritubular capillary (PTC) confirmed by immunostaining. Although the Banff 2013 criteria for ABMR introduced C4d-negative ABMR [2], the immunohistochemical evaluation of C4d remains an indispensable part of ABMR assessment. The Banff 2017 criteria accepted C4d positivity as an alternative marker of DSA testing [3], indicating its importance.
C4d is an end product of the activated complement system [4]. Antibody-mediated immune reactions activate the classical complement pathway, followed by the degradation of activated C4. C4d, a fragment of C4, forms a covalent thioester bond with nearby proteins and remains in the tissue, while other immune products are washed away. Therefore, C4d deposition is considered a trace of the antibody reaction and histologic hallmark of ABMR. Indeed, many studies have reported that it has a significant association with DSA and poor graft survival [5, 6].
Currently, the evaluation of C4d deposition is based on a scoring system according to the Banff criteria [7] using immunohistochemistry (IHC) or immunofluorescence (IF) staining. C4d staining is considered positive only when it is found in PTCs and vasa recta with a linear, circumferential staining pattern, regardless of the staining intensity. C4d staining findings in other areas, such as glomerular capillaries, tubular epithelium, and arteries as well as PTCs with incomplete patterns are regarded as false-positive results. False-negative results of C4d are also possible. An IHC slide containing only a single C4d-stained PTC should be interpreted as positive, but pathologists easily miss this finding. Thus, the interpretation of C4d is not only a tedious and laborious task but also a very difficult task with significant intra- and interobserver variabilities. Mengel et al. reported that C4d staining is subject to variation between observers and laboratories, with kappa values under 0.5 [8].
Recent advances in digital slide scanners and computer science have led to the development of image analysis techniques for digitized pathologic images. Machine learning algorithms with deep learning make it possible to analyze complicated tasks, such as the detection of metastatic tumors, prediction of prognosis, and grading of tumors [9,10,11,12,13]. In particular, computer-aided pathologic analysis has strengths in quantification tasks, and it could provide accurate and reproducible results with superior sensitivity and reduced labor and interobserver variability [14].
Because the scoring system of the Banff criteria has a quantitative nature, this could be an ideal target for digital image analysis. Among various scores, we targeted the C4d score and reported an automated algorithm for it [15]. The algorithm showed reasonable performance for detecting individual PTCs and identifying them as C4d-positive or -negative. In this study, the slide-level diagnostic performance of the algorithm was compared with those of the pathologists. Moreover, we collected additional C4d-immunostained slides from another institution, and the performance of the algorithm was validated with these slides, which were stained following different protocols and scanned using different slide scanners.
Methods
Patients selection
This study was performed using a protocol approved by the institutional review board of Asan Medical Center (AMC) (Approval no. 2018-0295). A total of 480 cases were randomly selected from all needle biopsies of renal allografts conducted from 2009 to 2016 from the anonymized research database of AMC. This database contains zero-day biopsies and indication biopsies with a clinical suspicion of graft dysfunction but does not include protocol biopsies. Of the 480 biopsies, 380 biopsies taken earlier were used for the development of the algorithm [15], and the last 100 biopsies were used for this study. To evaluate interinstitutional performance, an additional 86 cases were retrieved from Seoul National University Hospital (SNUH), which were randomly selected among biopsies conducted from 2017 to 2018. The database of SNUH contains zero-day biopsies, indication biopsies, and protocol biopsies, but all clinical information was blinded.
Data collection
Clinicopathological data collection
Cases from AMC provided the following information: scanned whole slide images (WSIs) of biopsy specimens, age at biopsy, elapsed time since kidney transplantation, pathologic diagnosis, and graft survival and follow-up periods. All clinical data were obtained from their anonymized research database, and no data that can lead to patient identification were collected. Cases from SNUH were submitted with anonymized scanned images of C4d IHC slides without clinical information.
Immunohistochemistry and WSI scanning
IHC for C4d was performed on 4 μm-thick sections from formalin-fixed, paraffin-embedded tissue blocks using a rabbit polyclonal antihuman C4d antibody (Cell Marque, Rocklin, CA, USA) at 1:32 dilution using a Ventana BenchMark XT autostainer (Ventana Medical Systems, Tucson, AZ, USA). All slides from AMC were scanned using a Pannoramic 250 Flash digital slide scanner (3DHISTECH, Budapest, Hungary) at ×20 magnification and a resolution of 0.221 μm per pixel.
Development of an automated algorithm
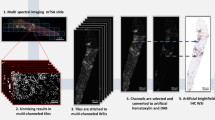
The detailed development processes were reported in our previously published article [15]. In brief, the algorithm comprises two steps: classification of the feasible regions of interest (ROIs) and detection of PTC. In the first step, the algorithm divided a WSI into numerous candidate feasible ROIs in 1024 × 1024 pixels and then distinguished feasible or non-feasible ROIs for interpretation using a classification model that was based on the Inception v3 network architecture. Areas with severe artifacts, poorly stained areas, glomeruli, and large vessels were automatically classified as non-feasible ROIs and were excluded from the next step. The second step detected and counted all PTCs with or without C4d staining separately in the feasible areas using a model that adopted the Faster R-CNN detection architecture. Finally, the algorithm outputs the number of entire PTCs and proportion of C4d-positive PTCs.
Evaluation of C4d immunohistochemistry
Slide review by pathologists
Three readers, including two board-certified renal pathologists (1 and 9 years of experience) and a 4th-year resident pathologist, participated in the study. The pathologists used the viewing software CaseViewer 2.1 (3DHISTECH, Budapest, Hungary) to evaluate WSIs with their personal computers and assessed the IHC according to the Banff 2017 criteria as follows: C4d0 (0%) = no staining; C4d1 (<10%) = minimal staining; C4d2 (10–50%) = focal staining; and C4d3 (≥50%) = diffuse staining. The results from individual pathologists were anonymized and designated as P scores (P1, P2, and P3). After completing the task, the pathologists had a review meeting and discussed their interpretations until a consensus score for discrepant cases was agreed upon, which was marked as the consensus score (C score).
C4d scoring by the algorithm and comparison with the results of the pathologists
The algorithm run using the on-premise system. The same computer used in the development system was used [15]. For each input, the algorithm automatically counted the number of C4d-stained and -unstained PTCs and calculated the proportion of C4d-stained PTCs. C4d scoring followed the Banff 2017 criteria, but we changed the criterion of C4d0 from 0% to <1% [15]. These scores were designated as the algorithm score (A score) (Fig. 1). To evaluate the agreement level among the pathologists, we compared A scores with P scores and C scores. Interobserver variability between pathologists and the algorithm was evaluated in three ways: at the full scale (C4d0-3), binary distinction of negative (C4d0) vs. positive (C4d ≥ 1), and no or minimal staining (C4d ≤ 1) vs. more-than-focal staining (C4d ≥ 2). This method was adopted from that of the Banff working group [8]. Thereafter, pathologists reviewed dozens of images produced by the algorithm, which annotated C4d-stained PTCs to analyze false-negative and false-positive errors.
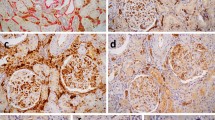
Examples from Case 433 (a–c) and Case 450 (d–f). Both capillaries showed linear C4d expression with moderate-to-strong intensity, but they are so small that they could be easily missed by pathologists. Annotation marks by the algorithm (c, f) help readers to recognize them. The number at the upper right side of the annotation box is a calculated probability of the prediction.
Interinstitutional validation
A total of 86 cases from SNUH were used to evaluate the performance of the algorithm with slides that were stained and scanned by different laboratories. IHC for SNUH cases was performed on 2 μm-thick sections from formalin-fixed, paraffin-embedded tissue blocks using a rabbit polyclonal antihuman C4d antibody (Cell Marque, Rocklin, CA, USA) at a 1:100 dilution by a Bond Max Automated IHC Vision Biosystem (Leica Microsystems, Wetzlar, Germany), and the stained slides were scanned by an Aperio AT2 scanner (Leica Biosystems, Newcastle upon Tyne, UK) at ×20 magnification and a resolution of 0.502 μm per pixel. Since the size of the ROI is defined as 1024 × 1024 pixels and is not based on the “actual” size, the slides scanned at different resolutions need resizing to fit in the algorithm. As SNUH slides were scanned at a lower resolution (0.502 μm per pixel) than AMC slides (0.221 μm per pixel), these images were magnified to match the actual size of each ROI. However, this caused a problem: images were blurred in this process and were classified into non-feasible areas in the first step. To overcome this issue, augmentation methods, such as sharpening, blurring, and zooming in and out, were used to improve the robustness of the algorithm. The C4d scores of SNUH slides were also reviewed by pathologists to make a C score using the viewing software CaseViewer 2.1.
Correlation with clinicopathological data
Assessment for histopathological diagnosis
For each of the 100 biopsies from AMC, all available glass slides were reviewed by an expert pathologist (HG). The diagnostic criteria followed the 2018 Banff reference guide [7]. Microvascular inflammation (MVI) was evaluated by the MVI score, which was defined as the sum of the g score (glomerulitis) and the ptc score (peritubular capillaritis). Intensity was classified as negative/mild (MVI < 2), moderate (2 ≤ MVI < 5), or severe (MVI ≥ 5).
Graft survival analysis
The graft survival period was calculated from the date the biopsy to the date of dialysis resumption, patient death, loss to follow-up, or end of the study (April 29th, 2019). Patient death despite a functioning graft and from other reasons was not counted as graft failure.
DSA assessment
Donor-specific anti-human leukocyte antigen (HLA) antibodies were assessed by the Luminex single antigen bead assay using LABScreen Single Antigen HLA Class I and Class II (One lambda, Canoga Park, CA, USA). The positive cut off value was a mean fluorescence intensity >1000.
Statistical analysis
All statistical analyses were calculated using the SPSS version 25.0 software (IBM, Armonk, NY, USA). Graft survival was compared by C4d scores using the Kaplan–Meier method and the log-rank test. Kappa (K) statistics were used to evaluate the agreement level between the P score and A score, P score and C score, and C score and A score. The correlation between the C4d score and histologic evidence of MVI was evaluated using the Fisher test. Statistical significance was set at p < 0.05.
Results
Population characteristics
The characteristics of the patients from AMC are summarized in Table 1. Their mean age was 48.9 ± 13.9 years. The mean elapsed time since the transplantation was 59.9 ± 76.5 months. There were 66 cases of pathologically diagnosed allograft rejection: 35 T cell-mediated rejection, 4 ABMR, and 27 mixed rejection cases. DSA was tested for in 79 patients and was found to be present in 22 patients. The median follow-up period after biopsy was 25.7 ± 8.8 months. In total, 22 graft losses occurred, including 21 patients who restarted dialysis and 1 patient who died with a functioning graft.
Interobserver concordance of the C4d score
All of the comparison data of the C4d scores are summarized in Table 2.
Comparison between the pathologists
All P scores, A scores, and C scores for 100 cases are provided in supplementary material 1. The consensus of the pathologists classified 52 cases as negative and 48 cases as positive for C4d with minimal (C4d1, 32 cases), focal (C4d2, 12 cases), and diffuse (C4d3, 4 cases) staining (Table 1). The interobserver variation between the three pathologists was moderate, and the average kappa value was 0.42. When binary distinction was performed with simplified C4d groups into positive and negative, the diagnostic concordance increased with a kappa value of 0.48. No and minimal staining (C4d0/1) group vs. more-than-focal staining (C4d2/3) group showed further improvement of reproducibility to a kappa value of 0.71. The three pathologists had C score accuracies of 77% (P1), 80% (P2), and 74% (P3), with kappa values of 0.63, 0.68, and 0.55, respectively. P1 and P2 showed excellent concordance (K = 0.93) with the C score when distinguishing more-than-focal staining (C4d ≥ 2) vs. no-to-minimal staining (C4d0/1), but P3 tended to give a lower score (i.e., gave a C4d1 score for C4d2- or 3-scored cases in the C score) and showed the lowest kappa values (0.56).
Comparison between the algorithm and pathologists
The C4d scores predicted by the algorithm achieved substantial concordance with the consensus score (K = 0.61) with an accuracy of 77%. The kappa values of the algorithm when compared with individual pathologists were lower at 0.34 (vs. P1), 0.42 (vs. P2), and 0.51 (vs. P3), which were comparable to the kappa values between the pathologists. The algorithm also showed a higher kappa value for binary distinction, with 0.72 for negative vs. positive staining and 0.70 for differentiating more-than-focal staining (C4d ≥ 2). On comparing C and A scores, A scores contained four false-positive and ten false-negative cases. The pathologists reviewed the output images from these cases and found several errors that brought about the false-positive and false-negative results (Fig. 2). The A score was also compared with each P score, assuming the C score as the reference standard. The algorithm found several false-negative cases wherein each pathologist erroneously scored a case as C4d0 when it was actually C4d-positive; this was true for 6 of 49 (12.2%) for P1, 3 of 44 (6.8%) for P2, and 7 of 58 (12.1%) for P3.
Examples of false-positive cases. a Linear, but not circumferential, expression should be regarded as negative according to the Banff guidelines. C4d expression in structures other than the peritubular capillary and vasa recta, such as the glomerular arteriole (b), loop of Henle (c), and glomerular capillary (d), should be considered a false-positive. Not all such cases were confused by the algorithm, as represented by the false-positive expression in the loop of Henle at the upper right side of (c), which was not predicted as a positive peritubular capillary. e–f Annotated images extracted from cases with C4d0 in the A score. These cases contain a few C4d-positive PTCs, and the algorithm did not fail to detect them, as seen in figures (e–f), but their proportion did not reach 0.5%. As we used altered criteria for C4d0 for A score (C4d < 1%), the algorithm graded them as C4d0. Although the algorithm misdiagnosed the C4d score, these annotated images were greatly helpful in the review by pathologists. A Algorithm, PTC peritubular capillary.
Comparison in the dataset from another institution
For the 86 cases from SNUH, the algorithm showed a kappa value of 0.45 for the C score. The kappa value for binary distinction was much higher when distinguishing more-than-focal staining from negative/minimal staining (K = 0.91) than when distinguishing negative from positive staining (K = 0.53). The scores predicted by the algorithm and the consensus panel are provided in supplementary material 2.
Clinicopathological correlation of the C4d score
All P scores, C scores, and A scores were significantly associated with MVI severity and the presence of DSA. Cases with higher C4d scores tended to reveal more severe MVI on histopathological examination, with statistical significance in both the C score (P = 0.0019) and A score (P = 0.0009), and DSAs were more frequently detected among patients with higher C4d scores (Table 3).
Graft failures occurred more frequently in the C4d-positive cases in both C and A scores (Fig. 3a). The mean survival time during the follow-up period was significantly shorter for C4d-positive cases than for C4d-negative cases, and it was statistically significant in terms of both A (p < 0.001) and C scores (p = 0.003) (Fig. 3b, c). When the patients were compared by C4d scores 0, 1, and ≥2, the p value remained less than 0.05 in terms of both A (P = 0.001) and C scores (P = 0.005). But, survival graphs crossed at the late follow-up period, precluding its validity (Fig. 3d, e).
Discussion
In our previous study, the proposed algorithm showed reasonable detection performance at the individual PTC level [15]. In this study, we evaluated the slide-level performance to assess its practical diagnostic applicability. The scoring result of the algorithm not only was comparable to that of the pathologists but also was significantly correlated with other evidence of ABMR, represented by the histological evidence of MVI, the presence of DSA, and overall graft survival.
In this study, each pathologist achieved moderate-to-substantial concordance with the C score, with kappa values ranging from 0.55 to 0.68. The algorithm also achieved substantial concordance with the C score (K = 0.61), which is similar to that of the pathologists. This result is better than the result achieved by the Banff working group [8]. They evaluated interobserver and interinstitutional variabilities in C4d scoring between a consensus panel and 78 pathologists. Consequently, their results showed moderate reproducibility, with an average kappa value of 0.44. Moreover, the clinical validity of the A score was further supported by its association with DSA, MVI, and graft survival.
Deep learning-based artificial intelligence can meticulously investigate an image with a high-level of consistency that is difficult for humans to achieve [16]. We focused on this property of the algorithm and tried to maximize the sensitivity in the range as it maintains reasonable accuracy. Indeed, the algorithm detected some positive PTCs that were missed by several pathologists; however, at the same time, it showed several false-positive detections in most cases (Fig. 2). Due to these false-positives, unfortunately, changing the criterion of C4d0 from 0% to <1% was inevitable to correct for false-positive errors at the slide level; however, it also caused some false-negative errors.
False-positive errors were mostly detected in tubule-like structures with thin walls, such as loops of Henle, atrophic tubules, glomerular capillaries, and arterioles (Fig. 2a–d). Some cases were also challenging for pathologists and required mutual discussion to consider them as false-positives. Despite using the altered criterion of C4d0 < 1%, four slide-level false-positive cases were seen. These cases mostly had a staining problem, wherein either too many nonspecific background signals confused the algorithm or too weak background hematoxylin staining precluded the detection of C4d-unstained PTCs and consequently overrated the proportion of C4d-positive PTCs. These results suggest that not only superior algorithmic performance but also excellent, laboratory-produced, high-quality slides are very important for applying computer-aided image analysis to digital pathology. Recent advances in quality-improving techniques of WSI, such as color normalization, could also help to solve these problems [17, 18].
False-negative results were absolutely critical as our aim was to maximize the sensitivity of the algorithm to enable its use as a screening tool. However, there were ten slide-level false-negative cases. To analyze this finding, pathologists reviewed dozens of images produced by the algorithm with annotations of C4d-stained PTCs and found that these results were mainly due to the altered criterion for C4d0. There was not a case that contained a C4d-positive PTC truly missed by the algorithm. The algorithm identified at least one output image with a C4d-positive PTC label in these cases (Fig. 2e, f), but its proportion did not reach 1% among all PTCs found in WSIs. Such cases, i.e., those containing less than five C4d-positive PTCs throughout WSIs, could also be missed by pathologists. Some of the slides were also classified as C4d0 during the pathologist review: two cases by P1, one case by P2, and four cases by P3. Although slide-level C4d0 classification by the algorithm was not perfectly reliable, it could function as a useful screening tool by providing annotations for some possible candidates of C4d-positive PTCs.
There are several limitations to discuss. First, the entire dataset used in the training and validation of the algorithm was retrieved from a single institution (AMC) [15]. We addressed this issue by testing the performance of the algorithm on cases from another institution (SNUH). Although deteriorated scoring concordance was noted at the full scale (C4d0-3), the algorithm achieved an excellent kappa value for binary distinction between C4d0-1 and C4d2-3. Most errors occurred when distinguishing between C4d0 and C4d1, as seen in the AMC dataset as well. Differences in laboratory settings and the slide scanner may affect the performance of the algorithm. Second, our dataset did not include protocol biopsy cases. Routine protocol biopsies have been implemented in many institutions [19]. There might be a smaller population expressing C4d among them compared with the population included in this study. If protocol biopsy cases had been included in the study population, the overall slide-level accuracy of the algorithm may have reduced because the algorithm made most errors in the distinction between C4d0 and C4d1. As discussed earlier, although the slide-level C4d0 classification of the algorithm was unreliable, the algorithm could function as a screening tool with annotations for possible candidates of C4d-positive PTCs. Third, there were some confounding factors within the clinicopathological correlation of C4d. C4d was negative in some chronic ABMR cases [2]. Conversely, there could be nonspecific C4d deposition in PTC, particularly in cases of ABO-incompatible transplantation [20]. In addition, the pathologists could not assure that the consensus result was absolutely accurate. There were some cases with uncertain C4d-stained structures that were very difficult to interpret as PTC. There was a possibility of misinterpretation with false-positive results and vice versa. These problems could potentially impair the reliability of C4d immunostaining itself. However, our data showed that both A and C scores were significantly associated with DSA, MVI, and graft survival, thus supporting their clinical validity. Although the algorithm itself had various false-negative and -positive errors and thereby could not act as a gold-standard C4d reader, we believe that it could be of valuable help to pathologists.
Deep learning-based image analysis has a great advantage in quantification [16, 18]. The Banff system for renal allograft biopsy is an ideal target because there are well-established diagnostic criteria based on semiquantitative evaluation. We selected the C4d score as the first step because the C4d score is based on IHC, which is unlike other scoring systems which are mainly based on histochemical staining, including hematoxylin–eosin (H&E), periodic acid-Schiff (PAS), trichrome, and methenamine silver staining (MT) [7]. Therefore, the stained slides have a sharp color contrast and relatively small variation in interinstitutional staining intensity. Furthermore, since C4d is a powerful prognostic marker [5, 6, 21] and is closely related to other histopathological and serological evidences of ABMR [20,21,22], further validation through clinicopathological correlation was easily achieved.
Notably, this study showed that a deep learning-based algorithm can detect and analyze complex histological architectures on IHC slides. This finding exceeded our expectations because IHC images have a disadvantage for recognizing complex architectures. Indeed, most deep learning-associated histopathological algorithms, particularly when detecting and analyzing complex histological architectures, use images stained with multiple colors such as H&E, PAS, and MT [11, 12, 23,24,25,26]. IHC uses only a single color (hematoxylin) to show the background architectures and has therefore been used in machine learning for quantifying relatively simple targets [27, 28]. The results of this study show a deep learning-based system’s ability for handling more complicated IHC interpretation tasks. For example, an assessment algorithm for immune cell infiltration in specific architectures, such as tubules, glomeruli, and vessels, using IHC markers, such as CD8, would improve the diagnostic accuracy and consistency. This achievement is not only greatly useful for practice but also very important for research because interobserver variability has been a major problem for renal transplant pathology [29].
In conclusion, the deep learning-based C4d detection algorithm showed a diagnostic performance similar to that of the pathologists in renal allograft biopsy. Scoring renal allograft biopsies to evaluate rejection, especially with the help of IHC, could be another suitable target for applying a deep learning-based decision support model.
References
Willicombe M, Roufosse C, Brookes P, Galliford JW, McLean AG, Dorling A, et al. Antibody-mediated rejection after alemtuzumab induction: incidence, risk factors, and predictors of poor outcome. Transplantation. 2011;92:176–82.
Haas M, Sis B, Racusen LC, Solez K, Glotz D, Colvin RB, et al. Banff 2013 meeting report: inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transpl. 2014;14:272–83.
Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, et al. The Banff 2017 Kidney Meeting Report: revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018;18:293–307.
Nickeleit V, Mihatsch MJ. Kidney transplants, antibodies and rejection: is C4d a magic marker? Nephrol Dial Transpl. 2003;18:2232–9.
Bohmig GA, Exner M, Habicht A, Schillinger M, Lang U, Kletzmayr J, et al. Capillary C4d deposition in kidney allografts: a specific marker of alloantibody-dependent graft injury. J Am Soc Nephrol. 2002;13:1091–9.
Mauiyyedi S, Crespo M, Collins AB, Schneeberger EE, Pascual MA, Saidman SL, et al. Acute humoral rejection in kidney transplantation: II. Morphology, immunopathology, and pathologic classification. J Am Soc Nephrol. 2002;13:779–87.
Roufosse C, Simmonds N, Clahsen-van Groningen M, Haas M, Henriksen KJ, Horsfield C, et al. A 2018 reference guide to the banff classification of renal allograft pathology. Transplantation. 2018;102:1795–814.
Mengel M, Chan S, Climenhaga J, Kushner YB, Regele H, Colvin RB, et al. Banff initiative for quality assurance in transplantation (BIFQUIT): reproducibility of C4d immunohistochemistry in kidney allografts. Am J Transpl. 2013;13:1235–45.
Acs B, Rimm DL. Not just digital pathology, intelligent digital pathology. JAMA Oncol.2018;4:403–4.
Arvaniti E, Fricker KS, Moret M, Rupp N, Hermanns T, Fankhauser C, et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep. 2018;8:12054.
Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210.
Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25:1301–9.
Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711.
Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inf. 2016;7:29.
Kim YG, Choi G, Go H, Cho Y, Lee H, Lee AR, et al. A fully automated system using a convolutional neural network to predict renal allograft rejection: extra-validation with giga-pixel immunostained slides. Sci Rep. 2019;9:5123.
Steiner DF, MacDonald R, Liu Y, Truszkowski P, Hipp JD, Gammage C, et al. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Pathol. 2018;42:1636–46.
Onder D, Zengin S, Sarioglu S. A review on color normalization and color deconvolution methods in histopathology. Appl Immunohistochem Mol Morphol. 2014;22:713–9.
Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20:e253–61.
Racusen LC. Protocol transplant biopsies in kidney allografts: why and when are they indicated? Clin J Am Soc Nephrol. 2006;1:144–7.
Haas M, Rahman MH, Racusen LC, Kraus ES, Bagnasco SM, Segev DL, et al. C4d and C3d staining in biopsies of ABO- and HLA-incompatible renal allografts: correlation with histologic findings. Am J Transpl. 2006;6:1829–40.
Einecke G, Sis B, Reeve J, Mengel M, Campbell PM, Hidalgo LG, et al. Antibody-mediated microcirculation injury is the major cause of late kidney transplant failure. Am J Transpl. 2009;9:2520–31.
Loupy A, Hill GS, Suberbielle C, Charron D, Anglicheau D, Zuber J, et al. Significance of C4d Banff scores in early protocol biopsies of kidney transplant recipients with preformed donor-specific antibodies (DSA). Am J Transpl. 2011;11:56–65.
Hermsen M, de Bel T, den Boer M, Steenbergen EJ, Kers J, Florquin S, et al. Deep learning-based histopathologic assessment of kidney tissue. J Am Soc Nephrol. 2019;30:1968–79.
Ginley B, Lutnick B, Jen KY, Fogo AB, Jain S, Rosenberg A, et al. Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol. 2019;30:1953–67.
Bukowy JD, Dayton A, Cloutier D, Manis AD, Staruschenko A, Lombard JH, et al. Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. J Am Soc Nephrol. 2018;29:2081–8.
Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054–6.
Steele KE, Tan TH, Korn R, Dacosta K, Brown C, Kuziora M, et al. Measuring multiple parameters of CD8+ tumor-infiltrating lymphocytes in human cancers by image analysis. J Immunother Cancer. 2018;6:20.
Sheikhzadeh F, Ward RK, van Niekerk D, Guillaud M. Automatic labeling of molecular biomarkers of immunohistochemistry images using fully convolutional networks. PLoS ONE. 2018;13:e0190783.
Furness PN, Taub N. Convergence of european renal transplant pathology assessment procedures P. International variation in the interpretation of renal transplant biopsies: report of the CERTPAP project. Kidney Int. 2001;60:1998–2012.
Acknowledgements
This work was supported by a grant (2018-7028) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Choi, G., Kim, YG., Cho, H. et al. Automated detection algorithm for C4d immunostaining showed comparable diagnostic performance to pathologists in renal allograft biopsy. Mod Pathol 33, 1626–1634 (2020). https://doi.org/10.1038/s41379-020-0529-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-020-0529-9
This article is cited by
-
Künstliche Intelligenz in der Nierentransplantationspathologie
Die Pathologie (2024)
-
Time for a full digital approach in nephropathology: a systematic review of current artificial intelligence applications and future directions
Journal of Nephrology (2023)
-
WARS1, TYMP and GBP1 display a distinctive microcirculation pattern by immunohistochemistry during antibody-mediated rejection in kidney transplantation
Scientific Reports (2022)