Abstract
The relative contribution of different microbial groups to soil organic matter (SOM) turnover and utilisation of rhizodeposits during a cropping season has remained largely unknown. We used a long-term field experiment (started in 1956), in which C3 crops were replaced with C4 silage maize in 2000, to investigate dynamics of fungi and bacterial groups and their utilisation of ‘young-C4’ and ‘old-C3’ SOM-derived resource every second week during the cropping season (June–Oct). Treatments include bare fallow, unfertilised, fertilised with mineral N and fertilised with farmyard manure (FYM) addition. Extracted soil phospholipid fatty acids (PLFAs) were pooled into Gram-positive, Gram-negative bacteria and fungi (18:2ω6,9) groups and their δ13C values determined. Total PLFAs amount correlated to the SOM contents (highest in FYM) and increased over the cropping season in N-fertilised and FYM treatments. As a result of a peak in plant growth during a period with frequent rain events in August, δ13C of total PLFAs significantly increased from − 23.8 to − 21.6‰ and − 26.1 to − 24.7‰, in N-fertilised and FYM addition, respectively. This clearly indicated a shift in microbial utilisation from old to young SOM sources, which was linked to increased soil moisture contents and fungal biomass. The abundance of Gram-positive increased and that of Gram-negative bacteria decreased until August and vice versa thereafter. The mean δ13C values of individual microbial groups were highest in fungi (corresponding to their seasonal biomass variation) followed by Gram-positive and Gram-negative bacteria. The results clearly demonstrated that irrespective of fertilisation type, fungi were the main players in seasonal SOM dynamics and were strongly influenced by soil moisture and phenological stage of the maize (i.e. rhizodeposition). Disentangling these microbial controls on C resources utilisation will be crucial for understanding C cycling during a cropping season or on an ecosystem scale.
Similar content being viewed by others
Introduction
Soil microorganisms act as a gatekeeper for soil–atmosphere C exchanges (CO2) by balancing the release and accumulation of soil organic matter (SOM). Plants transform atmospheric CO2 into organic matter, which enters the soil either as shoot and root residue inputs after plant death or as organic compounds released by living roots (exudates) referred to as rhizodeposition (Paterson et al. 1999; Jones et al. 2004). However, the intensity and amount of rhizodeposition is controlled by plant health and growth stage which in turn depends on soil management strategies and environmental factors such as moisture and temperature (Jangid et al. 2008). Management strategies (e.g. fertiliser inputs) also directly influence the microbial nutritional demand, supply and substrate utilisation pattern which govern soil nutrient cycling and subsequently SOM sequestration (Leifeld and Kögel-Knabner 2005; Shahbaz et al. 2017). Knowledge about the role in microbial transformation of rhizodeposits to SOM has remained unclear due to changing plant-microbial interactions over the cropping season e.g. caused by variations in substrate availability and moisture contents (Börjesson et al. 2016). Soil moisture is crucial in affecting microbial community composition (as bacteria is more sensitive than fungi to water stress) and microbial-mediated decomposition of organic substrates and the turnover of nutrients from inorganic fertilisers (Manzoni et al. 2012; Zhang et al. 2018). Better knowledge about the dynamics of microbial communities during a crop growing season (i.e. under changing rhizodeposition and soil moisture and temperature) is essential for understanding soil C balances impacted by plant-microbial interactions.
Several studies have been invoked to explain the behaviour of different microbial groups in exploitation of organic resources, and few have been empirical and quantitative in agricultural soils (Baraniya et al. 2018; Börjesson et al. 2016; Moore-Kucera and Dick 2008; Nannipieri et al. 2017). After addition to soil, organic substrates decompose to CO2 (through mineralisation) or become part of SOM (Cotrufo et al. 2013). The availability and composition of added substrates affect activity, biomass and composition of soil microbial communities (Griffiths et al. 1998; Heijboer et al. 2016). For instance, during the cropping season, microorganisms would experience more labile C input through root exudation whereas outside the cropping season, dead roots and available soil nutrients are important substrates to influence microbial growth and activity (Börjesson et al. 2016). Thus, response of microbial groups involves in SOM utilisation during low or high substrate availability is expected to change with crop growth stage and this need to be ascertained by continuous measurements.
It has been reported that various microbial groups could contribute differently in substrate utilisation (depending on substrate nature) and SOM turnover (Ghafoor et al. 2017; Pelz et al. 2005; Treonis et al. 2004). Whilst Gram-negative bacteria typically are considered to be associated with the consumption of easily degradable substrates (Bird et al. 2011; Creamer et al. 2015), Gram-positive bacteria are more active in decomposition of recalcitrant SOM compounds (Waldrop and Firestone 2004; Kramer and Gleixner 2006; Bird et al. 2011), and fungi are considered to have extensive roles in SOM turnover due to their greater ability to depolymerise complex compounds through extracellular enzyme production (Blagodatskaya and Kuzyakov 2008; Banerjee et al. 2016; García-Orenes et al. 2013). The fungal contribution to the microbial biomass or respiration is highly variable (e.g. can vary between 35 and 76%) in different soil groups (Joergensen and Wichern 2008; Soudzilovskaia et al. 2015). A few reports have shown a quick transfer of newly produced rhizodeposit-C to fungal biomass and a subsequent ability to rapidly process easily available root C (Treonis et al. 2004; Denef et al. 2007). However, we still lack a thorough understanding of the role of microbial groups in different C resource (e.g. fresh or old) utilisation for better predictions of SOM turnover as effected by seasonal variation and fertilisation strategies during a cropping period.
We can extend the knowledge about the microbial groups involved in SOM cycling by partitioning them according to their preferential use of substrate sources, e.g. fresh/recently added versus old C pools (Börjesson et al. 2016; Ghafoor et al. 2017; Morriën et al. 2017). This can be achieved by source partitioning following the transition from C3 to C4 crops, e.g. by tracing the isotopic signal of young or recent substrate inputs from C4 crops and that from old C3-derived SOM resources (Balesdent et al. 1987; Pelz et al. 2005; Shahbaz et al. 2019). The source of substrates from young versus old SOM assimilated by microorganisms will be reflected by the isotopic C composition in microbial cells (Blagodatskaya et al. 2011; Werth and Kuzyakov 2010). Phospholipid fatty acid analysis (PLFAs) is a well-established technique that provides accurate estimates of soil microbial biomass (Frostegård and Bååth 1996). By measuring the isotopic composition of different individual PLFAs (Kuzyakov and Domanski 2000), the substrate sources on which the microorganisms are feeding on, e.g. young versus old SOM pools, can be estimated. This could enhance our understanding of the impact of microbial groups, feeding on different substrates during a cropping season, on SOM dynamics and their role in processes important to agriculture and the environment.
In a long-term Swedish field experiment (Ultuna, started 1956), C3 crops were replaced with C4-maize in 2000. The impact of crop growth stages (i.e. rhizodeposition) on the main microbial groups over a cropping season was studied by analysing δ13C incorporation in PLFAs biomarkers. In situ seasonal dynamics of main microbial groups and their assimilation of old (C3) and young (C4) SOM resources were assessed through changes in δ13C signature in PLFAs. The topsoil was sampled to a depth of 20 cm every second week during maize crop growth period in bare fallow, cropped without or with N fertilisation and cropped with farmyard manure (FYM) addition. The objective of this study was to investigate how various microbial groups were involved in the assimilation and turnover of SOM (with a focus on young and old C pools) during a maize cropping season (June–October). Accordingly, it was hypothesized that (i) differences in the composition of main microbial groups as determined by PLFAs analysis and their source (C3 versus C4) utilisation pattern are more dependent on crop growth stage and seasonal variation (i.e. soil moisture) rather than fertilisation type and (ii) fungi have a more important role than the bacterial groups, in modulating soil microbial responses. We focused on three functional groups of soil microorganisms, i.e. fungi, Gram-negative and Gram-positive bacteria, to reveal their contribution to the decomposition of SOM under different C input regimes, soil fertility and changing environmental conditions during one cropping season.
Materials and methods
Study site and experimental design
The long-term field experiment is located at Ultuna, Uppsala, Sweden (59′ 82° N, 17′ 65° E). The site has remained under agricultural use (predominately C3 crops) for at least 300 years. The experiment was initiated in 1956 to investigate the impact of various fertilisation strategies on soil fertility. From 1956 to 1999, the site was kept under C3 annual crops, mainly spring barley, oats, rape, beet, mustard and turnip. The C3 crops had an average δ13C signature of − 28.0 ± 0.0‰ (Menichetti et al. 2013). The δ13C signature of soil in 1999 was about − 26.6‰ (Gerzabek et al. 2001). In 2000, C3 crops were replaced by C4 silage maize (Zea mays L.; varieties ‘Loft’ 2000–2005, ‘Revolver’ 2006–2014 and ‘Yukon’ 2015–2017) having a root δ13C signature of − 12.3 ± 0.0‰ (Menichetti et al. 2013). It was therefore possible to estimate old (until 1999) and young C (after 2000) sources in microbial groups involved in SOM turnover. Soil tillage (with 0–20 cm depth) has been performed manually since 1956 (Kätterer et al. 2011). At the time of harvest, all aboveground plant biomass was removed and thus input from crop residues to the soil was only from maize roots. Here the following treatments were selected: (i) bare fallow (uncropped without N fertilisation), (ii) cropped without N fertilisation, (iii) cropped with N fertilisation at rates of 80 kg N ha−1 year−1 as calcium nitrate at the time of sowing and (iv) cropped with FYM addition at rates of 4 Mg C ha−1 every second year in spring. The FYM derived from C3 crops and therefore, it had ca. −28.3‰ δ13C values. An area outside the experiment was used as C3 reference site where C3-crops have been continuously grown every year. Each spring, all treatments including the bare fallow received equal annual amounts of 20 kg P and 35–38 kg K ha−1, as superphosphate and 35–38 kg K ha−1 as KCl. A more detailed description of the experiment and results for the first 35 years was presented by Kirchmann et al. (1994). Each treatment had four replicates (with randomized complete block design) and each replicate plot was 2 × 2 m. Originally, the plots were separated by wooden frames extending to a depth of 30 cm, which were replaced by steel frames in 2009. According to the shift from C3 to C4 vegetation, we define the SOM derived from maize as ‘young’ (i.e. C4 after 2000) and that derived from C3-crops grown prior to 1999 as ‘old’. We use the term ‘fertilised’ to indicate cropped treatments with N addition and ‘unfertilised’ for cropped treatment without N addition. Table 1 shows the selected general site properties at the time of sampling including forage yields of maize.
Soil and weather conditions during sampling
The parent material consists of post-glacial sediments (Gerzabek et al. 2001) and the soil is classified as a Eutric Cambisol (Kirchmann et al. 2004). The soil has a clay loam texture with 37% clay (mainly illite), 41% silt and 22% sand. Before the start of experiment in 1956, the soil contained 1.5% organic C and 0.17% N and was pH 6.6 (H2O). Since the establishment of experiment (in 1956), the long-term treatments were affecting soil pH (Table 1). However, it was assumed that the seasonal impact on soil pH is low compared with long-term treatments impact.
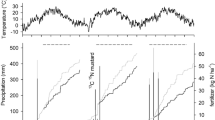
According to the Köppen classification (Peel et al. 2007), the site has a Dfb climate (warm summer hemi-boreal) with a mean annual air temperature of + 5.4 °C and mean annual precipitation of 570 mm. The weather reported from the Ultuna climate station (http://grodden.evp.slu.se/slu_klimat/), about 200 m from the trial, showed that during 2017, sunshine hours and temperature were close to annual average reported by Börjesson et al. (2016). Precipitation was 33 mm in June–July, 65 mm during August–September (the period of peak crop growth) and 90 mm in October. Therefore, due to less rain, the soil remained dry (close to wilting point) until mid-August, which also affected crop yields relative to a normal year. However, soil water contents remained approximately at or above 12% of total soil mass (Fig. 1; measured by gravimetric method as described by King (1981)). Soil temperature was measured with automatic loggers (Thermo Button 21G, progress plus, France), placed at 10 cm depth, which registered soil temperature every third hour. During the measurement campaign, mean soil temperature remained in the range of 14 to 24 °C during June to August and then declined to 5 °C in October (Fig. 1).
Crop growth and soil sampling
Maize was sown on 17th June 2017 and harvested as green silage on 29th September 2017. For obtaining a high temporal resolution under different C input regimes, the topsoil was sampled (0–20 cm) continuously during maize cropping period. The first soil sampling was performed on 15th June prior to sowing (PLFAs analysis were skipped for this point due to expected least difference to the sampling point after sowing, as shown for respiration data by Shahbaz et al. (2019)). After sowing, the soil was sampled every two weeks during the whole vegetation period including one additional sampling after harvest. In each plot, five soil samples per plot were taken to 20 cm depth, which were bulked together into one composite sample. Two samples were taken close to maize plants (at distance of about 5 cm) and three others randomly at the center between two plant rows. The soil was kept cold during sampling and immediately stored thereafter at − 20 °C for a couple of weeks. The soil was freeze-dried and milled before PLFAs analysis.
PLFAs extraction, analysis
The PLFA extraction procedure followed that described by Frostegård et al. (1993), which is based on the method of Bligh and Dyer (1959) as adapted by White et al. (1979). After lipid fractionation, the phospholipids were derivatised to fatty acid methyl esters (FAMEs) by adding methyl nonadecanoate fatty acid (19:0) as an internal standard.
The quantification and isotopic composition of individual FAMEs was determined using a GC Trace Ultra with combustion column attached via GC Combustion III to a Delta V Advantage isotope ratio mass spectrometer (all Thermo Finnigan, Bremen, Germany). The column, running conditions and quantification using the mass spectrometer data are described in Thornton et al. (2011). Initial data processing was performed using the instruments Isodat 3.0 Gas Isotope Ratio MS Software (Ver 3.0) (ThermoFisher Scientific, Bremen, Germany). These results were exported into Excel, and further processed using in-house Visual Basic macros, which helped in selection of peaks of interest using relative retention time with respect to the C19:0 internal standard. The δ13C values of the FAMES were normalised to the international standard of Vienna Pee Dee Belemnite (VPDB) scale using USGS72 icosanoic acid methyl ester (C20 FAME, δ13CVPDB = − 1.54, Reston Stable Isotope Laboratory) and C16:0 #1 hexadecenoic acid (C16 FAME, δ13CVPDB = − 30.74, Schimmelmann laboratory, Indiana University) which were run in duplicate at the beginning and end of every batch of samples. Following this, the δ13C of the C added as a methyl group during derivatisation was adjusted for using a mass balance approach. The C19:0 internal standard added also served as a quality control for δ13C values; long-term monitoring over several months gave a δ13C value of − 30.9 ± 0.99 (mean ± SD, n = 75).
Stable isotopic composition of all 13C-PLFAs data is expressed as delta notation (δ) in parts per thousand (‰) of a sample’s isotope concentration from the VPDB:
where R is the ratio of 13C to 12C in the samples or standard-VPDB. The δ13C of PLFAs is influenced not only by the isotopic signature of the source on which microorganisms feed but also by fractionation during microbial metabolism (Hayes 2001; Cifuentes and Salata 2001). The bare fallow treatment had not received any C inputs, which would have affected the isotopic spectrum of SOM, since 1956. The difference between the 13C signature of the source (i.e. SOM; see Shahbaz et al. 2019) and that of total PLFAs at each sampling date in the bare fallow can be assumed to reflect the fractionation factor arising from microbial metabolism (Börjesson et al. 2016). This enabled us to use the bare fallow treatment to calculate a fractionation factor that would express the isotopic change arising from metabolic fractionation. This factor was then used to estimate the maize-derived 13C that contributed to each PLFAs class over the maize cropping season. Further details about the calculation and fractionation factor are presented in Börjesson et al. (2016).
Composition of main microbial groups
A total number of 25 PLFAs biomarkers were used to calculate total PLFAs contents (μg C g−1 or nano mole esters g−1 soil) and to study the composition of soil microbial groups. An explanation of the nomenclature of different PLFAs biomarker is given in the supplementary material (SM) (Fig. SM1).
PLFAs biomarkers were used to characterise different microbial groups. To avoid misinterpretation three most common and well accepted PLFAs groups were used to categorise the major groups of microorganisms (Williams et al. 2013; Börjesson et al. 2016). PLFAs with one double bond represent Gram-negative bacteria (16:1ω7c, 18:1ω7, 17:1ω8c, cy17:0, cy19:0), branched PLFAs are generally attributed to Gram-positive bacteria (i15:0, a15:0, i16:0 and a17:0), and 18:2ω6,9 is regarded as a biomarker for fungi (Frostegård and Bååth 1996; Kramer and Gleixner 2006). All the other PLFAs were considered as general in this study. The concentration (mass) of each individual PLFAs biomarker and their δ13C values are presented in Fig. SM1.
Statistical analysis
The values presented in the figures and tables are given as means ± standard errors (SE; n = 4). Differences between the treatments were tested by post hoc (Duncan’s multiple range) tests after analysis of variance (ANOVA) with time as a repeated factor. The ANOVA was based on treatments (fertilisation strategies), sampling date and interaction between treatment and sampling date. Results different at P < 0.05 level were considered significant. ANOVA tables of all data presented in figures along with P values are given in SM. Figures and statistical analyses were performed using SigmaPlot (version 12.5; Systat Software Inc., UK). Correlations between the data were analysed by Person’s correlation method after checking assumptions i.e. level of measurement, related pairs, absence of outliers and linearity.
Results
Total PLFAs and δ13C values
There was a strong correlation (r = 0.93; Table 2) between the SOM contents and total PLFAs concentration (nmol fatty acid esters g−1 soil). The total PLFAs mass was about 2–2.5 times higher in soil samples from plots with FYM addition (70–106 nmol fatty acid esters g−1) compared with samples from the fertilised and unfertilised treatments, respectively (Fig. 2). Except for bare fallow, the total mass of PLFAs increased during the cropping season, particularly after August in the FYM treatment (Fig. 2).
The δ13C incorporation in total PLFAs corresponded to the fertilisation strategy. Since the origin of the FYM was C3 sources, δ13C of PLFAs were more depleted in the FYM treatment at all sampling occasions compared with mineral N-fertilised and unfertilised soils (Fig. 2). The cropping season had a profound effect on δ13C values in total PLFAs within treatments: A significant increase in δ13C was observed in FYM (from − 26.1 to − 24.7‰) and mineral N-fertilised plots (− 23.8 to − 21.6‰), particularly after August (peak in the cropping season), which suggested a shift in microbial utilisation from old (C3) to young (C4) SOM resources (Fig. 2). In contrast, the δ13C values of PLFAs in unfertilised plots significantly decreased from − 23.7 to − 24.7‰ after August, indicating a shift in the microbial metabolism towards old SOM resources. However, this change in δ13C was reversed in October.
Microbial groups and δ13C abundance
On the base of 25 identified PLFAs biomarkers, the microbial communities were grouped into fungi (18:2ω6,9), Gram-positive or Gram-negative bacteria and to general or unspecific (Fig. 3; Fig. SM1). Analogous to total PLFAs, the fungal biomass also related to the SOM contents. The fungal biomass was highest (71–175 μg C g−1 soil) under FYM followed by fertilised and unfertilised treatments, respectively, with an evident increase over the cropping season (Fig. 3). After August fungal biomass significantly increased from 54 to 115 μg C g−1 soil under mineral N-fertilisation compared with unfertilised plots (65–74 μg C g−1 soil).
Fungal biomass under different long-term management practices over a maize cropping season. Maize was sown on 17th June and harvested as green silage on 29th September. Error bar represents ± SE (n = 4). Fungal biomass is shown in μg C g−1 soil, which is obtained after multiplying nmol PLFAs g−1 soil of biomarker (18:2휔6,9) by a factor of 5.8 (Joergensen and Emmerling 2006)
In contrast to fungi, the contents of total bacterial biomarkers (Gram-positive plus Gram-negative) increased over the vegetation season until August and declined thereafter (Fig. 4). The contribution of Gram-negative and Gram-positive bacteria to total bacterial biomarkers was season-specific. Except for the mid-cropping season (August) the contribution of Gram-negative bacteria to total bacterial biomarkers remained higher than that of Gram-positive bacteria; with highest abundance in FYM plots followed by fertilised and unfertilised plots (Fig. 4). However, irrespective of fertilisation treatment a seasonal trade-off was observed between Gram-positive and Gram-negative bacteria. The contents of Gram-positives bacterial biomarkers increased and that of Gram-negatives decreased until August, thereafter the opposite trend was observed (Fig. 4). The bacteria to total PLFAs ratio increased until August and declined thereafter in all treatments (Fig. SM2). In contrast, fungal to total PLFAs ratio remained stable in all treatments until August and then increased sharply under the mineral N-fertilisation and FYM additions (Fig. SM2).
Except in the bare fallow treatment, Gram-negative bacteria always had significantly lower average δ13C values than fungi and Gram-positive bacteria (Fig. 5). No significant difference was detected in δ13C values between Gram-positives and fungi in soil samples from unfertilised plots. Over the cropping season, fungi always had significantly higher δ13C values than Gram-positive bacteria in the FYM treatment. A similar difference in δ13C in PLFAs under mineral N-fertilisation was only observed after mid-August (Fig. 5). A seasonal increase in δ13C values of fungi and Gram-positive bacteria was observed after August in N-fertilised and FYM treatments. The δ13C values of microbial groups remained constant in bare fallow throughout the sampling period and remained constant during the cropping season in the unfertilised treatment (Fig. 5).
Mean δ13C values of phospholipid fatty acids (PLFAs) biomarkers of Gram-positive, Gram-negative bacteria and fungi under different long-term fertilisation strategies over a maize cropping season. Maize was sown on 17th June and harvested as green silage on 29th September. Error bars represent ± SE (n = 4)
Discussion
PLFAs and microbial groups
The results demonstrate how cropping season and changes in soil moisture affect the growth, abundance and substrate utilisation pattern of soil fungi and bacterial groups. Total PLFAs (reflecting total microbial biomass) were strongly correlated to the SOM and soil moisture contents, as has been previously reported for the same site (Börjesson et al. 2016). The soil microbial abundance, as measured by total PLFAs, increased during the cropping season (profoundly under FYM), which contrasted earlier observations (e.g. Börjesson et al. 2016; Hamer et al. 2008; Jangid et al. 2008). The increase in total PLFAs was prominent after August and coincided with an increase in 13C in PLFAs (discussed below), which was potentially due to two main reasons: (i) an increase in soil moisture contents due to frequent rain events during or after August (Fig. 1) and (ii) as a consequence of an increase in substrate availability and rhizodeposition due to accelerated plant growth (i.e. enriched δ13C young C4-resources availability, e.g. Dercon et al. (2006)).
Microbial communities are known to respond dynamically to available nutrients, moisture and plant-microbe interactions (Baraniya et al. 2018; Sasse et al. 2017; Hu et al. 2018; Zhang et al. 2018). High rhizodeposition is generally believed to favor Gram-negative bacteria, which should be seen as an increase of in mono-unsaturated PLFAs (Treonis et al. 2004; Kramer and Gleixner 2006; Tavi et al. 2013). However, results show the opposite trend, with an increase of the ratio of fungal:total PLFAs and a decrease of the bacterial (cumulative of Gram-negative and Gram-positive): total PLFAs ratio after August (Fig. SM2). This means that the increase in substrate availability after the rain events (or microbial stress alleviation) favored either the fungal growth, with up to 110% increase in fungi biomass, or fungi outcompeted bacteria to benefit from rhizodeposition (Butler et al. 2003; Treonis et al. 2004). Moreover, fungi are usually considered to be less affected than bacteria during competition with plant roots (e.g. by making direct association with roots through mycorrhizae) for nutrients and water from soil (Jones et al. 2004; Fontaine et al. 2011; Hu et al. 2018). Although bacterial to total PLFAs ratio declined after August (in contrast to fungi), a seasonal trade-off was observed for Gram-positive and Gram-negative bacterial abundance during maize cropping. Due to less rain, the soil was relatively dry during early growth periods as compared with the period after August (Fig. 1). Table 2 clearly demonstrates that abundance of fungi (r = 0.72) and Gram-negative bacteria (r = 0.60) were more strongly correlated with soil moisture than Gram-positive bacterial abundance (r = 0.40). Accordingly, the contents of Gram-positive bacterial biomarkers increased until August, indicating their higher ability to endure stress under low moisture or lower rhizodeposition conditions (Williams 2007). Soil moisture availability has also been shown to directly or indirectly impact soil microbial community composition (Zhang et al. 2018). In general, certain Gram-positive bacteria (e.g. Actinobacteria and Firmicutes) are considered more resistant to drought stress than the Gram-negative bacteria (Chodak et al. 2015). The increase in abundance of Gram-negative bacteria (in all fertilisation strategies) after August could mainly be linked to the high soil moisture or increased rhizodeposition, which enabled them to outcompete Gram-positive bacteria in resource utilisation. This increase in Gram-negative was mainly connected to 16:1ω7c and 18:1ω7 biomarkers, behaving as expected (Treonis et al. 2004; Kramer and Gleixner 2006; Tavi et al. 2013) whilst in contrast 19:0cy and 17:0cy remained more constant through the season (Fig. SM1). However, the fungal abundance was generally higher than the bacterial abundance (in contrast to their abundance before August, i.e. in low moisture), which indicates that fungi may have a more important role in C cycling.
δ13C in total PLFAs and microbial groups
Following exclusively C3 vegetation, maize has been grown at the trial site since 2000, which resulted in establishment of δ13C gradient in SOM (Shahbaz et al. 2019). The δ13C values in total PLFAs or in PLFAs within each microbial groups were compared as criteria to investigate the impact of seasonal changes on substrate availability from SOM (i.e. old-C3 and young-C4 pools or rhizodeposition) and on the composition of main microbial groups. Similar to bare fallow, the enrichment of δ13C in total PLFAs did not increase (even declined after August) over the vegetation period in plots without fertilisation (Fig. 2). The microorganisms could have become limited in energy or nutrients during the cropping period due to competition with roots that could force the microbial community to shift towards older (C3) SOM sources (Blagodatskaya and Kuzyakov 2008). This decrease (or no-increase of δ13C) may also reflect the N limitation to microorganisms likely bacteria, involved in N turnover (Petersen et al. 2004). Correspondingly, the observed decrease in 13C in total PLFAs was associated with an increase in abundance of Gram-negative bacteria and a decline in their δ13C (Figs. 4 and 6, unfertilised).
A conceptual illustration of rhizodeposition under stress and relief conditions affecting plant growth and microbial communities functioning. Under stress (left side), plant growth is limited by soil moisture and thus less substrate is available to microbes. Water stress resulted in community shifts favoring bacteria (particularly G+ bacteria). Under relief conditions (right side), the total biomass increased with higher abundance of fungi than bacteria. The gradient color fill intensity within individual parts of the pie charts illustrates the microbial substrate utilisation pattern of old (C3) versus young (C4) SOM resources (as evident through 훿13C values). We propose that the microbial biomass at any specific time period reflects the amount of substrates accessible by the microbes and the amount of microbial derived SOM (e.g. necromass, as explained by Cotrufo et al. 2013)
In contrast, δ13C in PLFAs increased under fertilisation especially after August (concurring to total PLFAs amount), both in mineral N and in FYM treatments. This indicated that microbial utilisation of young SOM resources (C4) and/or rhizodeposition increased over the cropping season. It is remarkable that the increase in total PLFAs and their δ13C values coincided with an increase of fungal biomass (18:2ω6,9) and its increasing δ13C value, which indicated dominance of the fungal community in exploitation of young SOM resources. Fungi can use recalcitrant old SOM over short periods of time (Paterson et al. 2008; Fontaine et al. 2011); however, this was not evident in our case. Accordingly, Fig. 5 clearly demonstrates that fungi had the highest δ13C values compared with Gram-negative and Gram-positive bacteria. This was more pronounced under fertilisation with mineral N or FYM with a seasonal increase during the cropping period. This also means that fungi responded rapidly and more dominantly (than bacteria) to the changes in substrate availability, i.e. due to rhizodeposition or rain events after August. Similarly, there is evidence that changes in SOM quality, caused for examples by manure application or rhizodeposition, can increase the microbial biomass and induce shifts in their community composition leading to increases in fungal abundance (Balasooriya et al. 2013; García-Orenes et al. 2013; Zhang et al. 2018). Some studies report that Gram-negative bacteria respond quickly to substrate availability or root exudates (Butler et al. 2003; Koranda et al. 2011). In our study, abundance of Gram-negative bacteria increased over the cropping season but δ13C remained relatively stable or actually declined in unfertilised plots. The Gram-negative bacteria retained the lowest δ13C values (Fig. 5), with minimal changes over the cropping season, demonstrating that they were associated mainly with old (C3) SOM resources. In the bare fallow (without substrate addition since 1956), fungi and Gram-negative bacteria shared a similar δ13C signature whilst Gram-positive values were more enriched. Overall the results show that the microbial groups used different strategies to grow and to feed on C sources, which depends on nutrients availability (e.g. fertilisation strategy) and cropping season i.e. rhizodeposition.
Implications of quantitative source estimation in PLFAs
The contribution of C4-derived C in the weighted average of PLFAs should follow the trend of its δ13C values. In the previous work, the contribution of C4 derived C in the microbial biomass (on the base of PLFAs biomarkers), after vegetation shift, has been reported to 10–30% (Menichetti et al. 2013; Novara et al. 2014; Börjesson et al. 2016). Two years following a shift from C3 to C4 vegetation, Kramer et al. (2012) estimated that maize-derived C can contribute to 25% in total microbial biomass. Our study reports values of C4 usage in PLFAs in a similar range (ca. 13–40% with seasonal variations (Fig. SM3)), albeit somewhat larger as expected given the longer time period since the shift from C3 to C4. However, we assume that quantitative estimation of young versus old SOM-derived C in PLFAs groups by using two-source mixing model could be suspect due to multisource inputs (e.g. in the FYM treatment in this study) or if potential changes in δ13C fractionation during decomposition remains unconsidered. Under natural abundance, slow changes in δ13C signatures occur due to either isotopic fractionations within microbial cells during metabolism or microbial preferential substrate utilisation or accumulation of 13C-enriched microbial necromass (Hayes 2001; Cifuentes and Salata 2001; Werth and Kuzyakov 2010; Blagodatskaya et al. 2011). The magnitude of the metabolic fractionation has been debated in the literature (e.g. Blagodatskaya et al. 2011; Schneckenberger and Kuzyakov 2007); experimental evidence suggests that this effect is real and needs to be considered in calculations. Some studies have considered this by assuming a fixed fractionation factor (Menichetti et al. 2015; Börjesson et al. 2016). However, this factor may vary between treatments and over the growing season due to changes in substrate availability (i.e. rhizodeposition) and microbial diversity (Cifuentes and Salata 2001; Ghafoor et al. 2017; Novara et al. 2014; Yang et al. 2014). Therefore, both fractionation during metabolism and preferential microbial utilisation may impact independently on resource (e.g. C3 versus C4) estimations in PLFAs. To avoid such artefacts, it is necessary to consider all the possible factors involved in δ13C variations during calculation for proper estimation of the amount of young (e.g. C4-derived) or old (e.g. C3-derived) C that contributed to each PLFAs class over the growing seasons. For the estimation of δ13C in PLFAs, we have considered the fractionation factor separately for each individual sampling point by following Börjesson et al. (2016). However, Fig. 6 illustrates the resource utilisation pattern (proportion of C4-young versus C3-old C) by microbial groups on the base of microbial abundance and changes in their δ13C values over the maize cropping period.
Conclusions
This study establishes that fertilisation strategy, maize growth stage and seasonal variation (especially soil moisture content) have a strong impact on shaping the composition of main microbial groups and their ability to utilise SOM resources (as identified through 13C values of PLFAs and following a switch from C3 to more 13C enriched C4 vegetation). The total PLFAs amount (indicating biomass) increased over the cropping season which coincided with an increase of 13C in PLFAs under fertilisation, with both calcium nitrate and FYM. At the time of early growth period, bacteria (with ability to endure low moisture stress) share the major contribution in total PLFAs contents. During high substrate availability periods (i.e. after August with frequent rain events), fungi were the most dominant microbial group which outcompeted bacteria in utilizing young (rhizodeposition) SOM resources. This was also confirmed by the highest δ13C values in fungi followed by Gram-positive and Gram-negative bacteria. Irrespective to fertilisation strategy, a seasonal trade-off was observed between Gram-positive and Gram-negative bacteria. The contents of Gram-positive biomarkers increased and that of Gram-negative decreased until August, and vice versa thereafter. Our results emphasise the importance of soil moisture and cropping growth stage in modulating plant-microbial interactions and reveal the pivotal role of fungi versus bacteria in SOM turnover during periods with high rhizodeposition. These findings also provide insights on how microbial groups and their interaction control SOM resource utilisation and C cycling under changing biotic and abiotic conditions.
References
Balasooriya WK, Huygens D, Denef K, Roobroeck D, Verhoest NEC, Boeckx P (2013) Temporal variation of rhizodeposit-C assimilating microbial communities in a natural wetland. Biol Fertil Soils 49:333–341
Balesdent J, Mariotti A, Guillet B (1987) Natural 13C abundance as a tracer for studies of soil organic matter dynamics. Soil Biol Biochem 19:25–30
Banerjee S, Kirkby CA, Schmutter D, Bissett A, Kirkegaard JA, Richardson AE (2016) Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol Biochem 97:188–198
Baraniya D, Nannipieri P, Kublik S, Vestergaard G, Schloter M, Schöler A (2018) The impact of the diurnal cycle on the microbial transcriptome in the rhizosphere of barley. Microb Ecol 75:830–833
Bird JA, Herman DJ, Firestone MK (2011) Rhizosphere priming of soil organic matter by bacterial groups in a grassland soil. Soil Biol Biochem 43:718–725
Blagodatskaya E, Kuzyakov Y (2008) Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: critical review. Biol Fertil Soils 45:115–131
Blagodatskaya E, Yuyukina T, Blagodatsky S, Kuzyakov Y (2011) Turnover of soil organic matter and of microbial biomass under C3-C4 vegetation change: consideration of 13C fractionation and preferential substrate utilization. Soil Biol Biochem 43:159–166
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Physiol Pharmacol 37:911–917
Börjesson G, Menichetti L, Thornton B, Campbell CD, Kätterer T (2016) Seasonal dynamics of the soil microbial community: assimilation of old and young carbon sources in a long-term field experiment as revealed by natural 13C abundance. Eur J Soil Sci 67:79–89
Butler JL, Williams MA, Bottomley PJ, Myrold DD (2003) Microbial community dynamics associated with rhizosphere carbon flow. Appl Environ Microbiol 69:6793–6800
Chodak M, Gołębiewski M, Morawska-Płoskonka J, Kuduk K, Niklińska M (2015) Soil chemical properties affect the reaction of forest soil bacteria to drought and rewetting stress. Ann Microbiol 65:627–1637
Cifuentes LA, Salata GG (2001) Significance of carbon isotope discrimination between bulk carbon and extracted phospholipid fatty acids in selected terrestrial and marine environments. Org Geochem 32:613–621
Cotrufo MF, Wallenstein MD, Boot CM, Denef K, Paul E (2013) The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: do labile plant inputs form stable soil organic matter? Glob Chang Biol 19:988–995
Creamer CA, de Menezes AB, Krull ES, Sanderman J, Newton-Walters R, Farrell M (2015) Microbial community structure mediates response of soil C decomposition to litter addition and warming. Soil Biol Biochem 80:175–188
Denef K, Bubenheim H, Lenhart K, Vermeulen J, Van Cleemput O, Boeckx P, Müller C (2007) Community shifts and carbon translocation within metabolically-active rhizosphere microorganisms in grasslands under elevated CO2. Biogeosciences 4:769–779
Dercon G, Clymans E, Diels J, Merckx R, Deckers J (2006) Differential 13C isotopic discrimination in maize at varying water stress and at low to high nitrogen availability. Plant Soil 282:313–326
Fontaine S, Henault C, Aamor A, Bdiouia N, Bloora JMG, Maire V, Mary B, Revaillot S, Maron PA (2011) Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect. Soil Biol Biochem 43:86–96
Frostegård Å, Bååth E (1996) The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fertil Soils 22:59–65
Frostegård Å, Bååth E, Tunlid A (1993) Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis. Soil Biol Biochem 25:723–730
García-Orenes F, Morugán-Coronado A, Zornoza R, Scow K (2013) Changes in soil microbial community structure influenced by agricultural management practices in a Mediterranean agro-ecosystem. PLoS One 8:e80522
Gerzabek MH, Haberhauer G, Kirchmann H (2001) Soil organic matter pools and carbon-13 natural abundances in particle-size fractions of a long-term agricultural field experiment receiving organic amendments. Soil Sci Soc Am J 65:352–358
Ghafoor A, Poeplau C, Kätterer T (2017) Fate of straw- and root-derived carbon in a Swedish agricultural soil. Biol Fertil Soils 53:257–267
Griffiths B, Ritz K, Ebblewhite N, Dobson G (1998) Soil microbial community structure: effects of substrate loading rates. Soil Biol Biochem 31:145–153
Hamer U, Makeschin F, Stadler J, Klotz S (2008) Soil organic matter and microbial community structure in set-aside and intensively managed arable soils in NE-Saxony, Germany. Appl Soil Ecol 40:465–475
Hayes JM (2001) Fractionation of carbon and hydrogen isotopes in biosynthetic processes. Rev Mineral Geochem 43:225–277
Heijboer A, ten Berge HFM, de Ruiter PC, Jørgensen HB, Kowalchuk GA, Bloem J (2016) Plant biomass, soil microbial community structure and nitrogen cycling under different organic amendment regimes; a 15N tracer-based approach. Appl Soil Ecol 107:251–260
Hu L, Robert CAM, Cadot S, Zhang X, Ye M, Li B, Manzo D, Chervet N, Steinger T, van der Heijden MGA, Schlaeppi K, Erb M (2018) Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9:1–13
Jangid K, Williams MA, Franzluebbers AJ, Sanderlin JS, Reeves JH, Jenkins MB, Endale DM, Coleman DC, Whitman WB (2008) Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol Biochem 40:2843–2853
Joergensen RG, Emmerling C (2006) Methods for evaluating human impact on soil microorganisms based on their activity, biomass, and diversity in agricultural soils. J Plant Nutr Soil Sci 169:295–309
Joergensen RG, Wichern F (2008) Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol Biochem 40:2977–2991
Jones DL, Hodge A, Kuzyakov Y (2004) Plant and mycorrhizal regulation of rhizodeposition. New Phytol 163:459–480
Kätterer T, Bolinder MA, Andrén O, Kirchmann H, Menichetti L (2011) Roots contribute more to refractory soil organic matter than above-ground crop residues, as revealed by a long-term field experiment. Agric Ecosyst Environ 141:184–192
King PM (1981) Comparison of methods for measuring severity of water repellence of Sandy soils and assessment of some factors that affect its measurement. Aust J Soil Res 19:275–285
Kirchmann H, Persson J, Carlgren K (1994) The Ultuna long-term soil organic matter experiment, 1956–1991. Department of Soil Sciences, Reports and Dissertations 17. Swedish University of Agricultural Sciences, Uppsala, Sweden
Kirchmann H, Haberhauer G, Kandeler E, Sessitsch A, Gerzabek MH (2004) Effects of level and quality of organic matter input on carbon storage and biological activity in soil: synthesis of a long-term experiment. Glob Biogeochem Cycles 18:1–9
Koranda M, Schnecker J, Kaiser C, Fuchslueger L, Kitzler B, Stange CF, Sessitsch A, Zechmeister-Boltenstern S, Richter A (2011) Microbial processes and community composition in the rhizosphere of European beech - the influence of plant C exudates. Soil Biol Biochem 43:551–558
Kramer C, Gleixner G (2006) Variable use of plant- and soil-derived carbon by microorganisms in agricultural soils. Soil Biol Biochem 38:3267–3278
Kramer S, Marhana S, Ruess L, Armbruster W, Butenschoend O, Haslwimmer H, Kuzyakov Y, Pausch J, Scheunemannd N, Schoene J, Schmalwasser A, Totsche KU, Walker F, Stefan S, Kandeler E (2012) Carbon flow into microbial and fungal biomass as a basis for the belowground food web of agroecosystems. Pedobiologia 55:111–119
Kuzyakov Y, Domanski G (2000) Carbon input by plants into the soil. J Plant Nutr Soil Sci 163:421–431
Leifeld J, Kögel-Knabner I (2005) Soil organic matter fractions as early indicators for carbon stock changes under different land-use? Geoderma 124:143–155
Manzoni S, Schimel JP, Porporato A (2012) Responses of soil microbial communities to water stress: results from a meta-analysis. Ecology 93:930–938
Menichetti L, Ekblad A, Kätterer T (2013) Organic amendments affect δ13C signature of soil respiration and soil organic C accumulation in a long-term field experiment in Sweden. Eur J Soil Sci 64:621–628
Menichetti L, Houot S, van Oort F, Kätterer T, Christensen BT, Chenu C, Barré P, Vasilyeva NA, Ekblad A (2015) Increase in soil stable carbon isotope ratio relates to loss of organic carbon: results from five long-term bare fallow experiments. Oecologia 177:811–821
Moore-Kucera J, Dick RP (2008) PLFA profiling of microbial community structure and seasonal shifts in soils of a Douglas-fir chronosequence. Microb Ecol 55:500–511
Morriën E, Hannula SE, Snoek LB, Helmsing NR, Zweers H, de Hollander M, Soto RL, Bouffaud ML, Buée M, Dimmers W, Duyts H, Geisen S, Girlanda M, Griffiths RI, Jørgensen HB, Jensen J, Plassart P, Redecker D, Schmelz RM, Schmidt O, Thomson BC, Tisserant E, Uroz S, Winding A, Bailey MJ, Bonkowski M, Faber JH, Martin F, Lemanceau P, de Boer W, van Veen JA, van der Putten WH (2017) Soil networks become more connected and take up more carbon as nature restoration progresses. Nat Commun 8:14349
Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G (2017) Microbial diversity and soil functions. Eur J Soil Sci 68:12–26
Novara A, La Mantia T, Rühl J, Badalucco L, Kuzyakov Y, Gristina L, Laudicina VA (2014) Dynamics of soil organic carbon pools after agricultural abandonment. Geoderma 235–236:191–198
Paterson E, Hodge A, Thornton B, Millard P, Killham K (1999) Carbon partitioning and rhizosphere C-flow in Lolium perenne as affected by CO 2 concentration, irradiance and below-ground conditions. Glob Chang Biol 5:669–678
Paterson E, Osler G, Dawson LA, Gebbing T, Sim A, Ord B (2008) Labile and recalcitrant plant fractions are utilised by distinct microbial communities in soil: independent of the presence of roots and mycorrhizal fungi. Soil Biol Biochem 40:1103–1113
Peel MC, Finlayson BL, Mcmahon TA (2007) Updated world map of the Köppen-Geiger climate classification. Hydrol Earth Syst Sci 11:1633–1644
Pelz O, Abraham W-R, Saurer M, Siegwolf R, Zeyer J (2005) Microbial assimilation of plant-derived carbon in soil traced by isotope analysis. Biol Fertil Soils 41:153–162
Petersen SO, Roslev P, Bol R (2004) Dynamics of a pasture soil microbial community after deposition of cattle urine amended with [13C]urea. Appl Environ Microbiol 70:6363–6369
Sasse J, Martinoia E, Northen T (2017) Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23:25–41
Schneckenberger K, Kuzyakov Y (2007) Carbon sequestration under Miscanthus in sandy and loamy soils estimated by natural 13C abundance. J Plant Nutr Soil Sci 170:538–542
Shahbaz M, Kuzyakov Y, Maqsood S, Wendland M, Heitkamp F (2017) Decadal nitrogen fertilization decreases mineral-associated and subsoil carbon: a 32-year study. Land Degrad Dev 28:1463–1472
Shahbaz M, Menichetti L, Kätterer T, Börjesson G (2019) Impact of long-term N fertilisation on CO2 evolution from old and young SOM pools measured during the maize cropping season. Sci Total Environ 658:1539–1548
Soudzilovskaia NA, van der Heijden MGA, Cornelissen JHC, Makarov MI, Onipchenko VG, Maslov MN, Akhmetzhanova AA, van Bodegom PM (2015) Quantitative assessment of the differential impacts of arbuscular and ectomycorrhiza on soil carbon cycling. New Phytol 208:280–293
Tavi NM, Martikainen PJ, Lokko K, Kontro M, Wild B, Richter A, Biasi C (2013) Linking microbial community structure and allocation of plant-derived carbon in an organic agricultural soil using 13CO2 pulse-chase labelling combined with C-PLFA profiling. Soil Biol Biochem 58:207–215
Thornton B, Zhang Z, Mayes RW, Högberg MN, Midwood AJ (2011) Can gas chromatography combustion isotope ratio mass spectrometry be used to quantify organic compound abundance? Rapid Commun Mass Spectrom 25:2433–2438
Treonis AM, Ostle NJ, Stott AW, Primrose R, Grayston SJ, Ineson P (2004) Identification of groups of metabolically-active rhizosphere microorganisms by stable isotope probing of PLFAs. Soil Biol Biochem 36:533–537
Waldrop MP, Firestone MK (2004) Microbial community utilization of recalcitrant and simple carbon compounds: impact of oak-woodland plant communities. Oecologia 138:275–284
Werth M, Kuzyakov Y (2010) 13C fractionation at the root–microorganisms–soil interface: a review and outlook for partitioning studies. Soil Biol Biochem 42:1372–1384
White DC, Davis WM, Nickels JS, King JD, Bobbie RJ (1979) Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia 40:51–62
Williams MA (2007) Response of microbial communities to water stress in irrigated and drought-prone tallgrass prairie soils. Soil Biol Biochem 39:2750–2757
Williams A, Börjesson G, Hedlund K (2013) The effects of 55 years of different inorganic fertiliser regimes on soil properties and microbial community composition. Soil Biol Biochem 67:41–46
Yang W, Magid J, Christensen S, Rønn R, Ambus P, Ekelund F (2014) Biological 12C–13C fractionation increases with increasing community-complexity in soil microcosms. Soil Biol Biochem 69:197–201
Zhang Y, Hao X, Alexander TW, Thomas BW, Shi X, Lupwayi NZ (2018) Long-term and legacy effects of manure application on soil microbial community composition. Biol Fertil Soils 54:269–283
Funding
Open access funding provided by Swedish University of Agricultural Sciences. The work was supported by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS; project 2016-01350). We are grateful to former colleagues for starting the valuable Ultuna long-term field experiment and for keeping it running with financial support from the Faculty. The James Hutton Institute received funding from the Rural and Environmental Science and Analytical Services Division (RESAS) of the Scottish Government.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 134 kb).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shahbaz, M., Kätterer, T., Thornton, B. et al. Dynamics of fungal and bacterial groups and their carbon sources during the growing season of maize in a long-term experiment. Biol Fertil Soils 56, 759–770 (2020). https://doi.org/10.1007/s00374-020-01454-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-020-01454-z