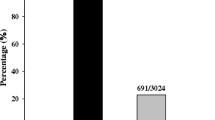
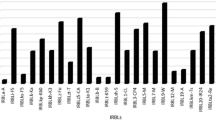
Abstract
Background
Rice blast is generally considered the most devastating rice disease worldwide. The development of resistant varieties has been proven to be the most economical strategy to control the disease. A cluster of resistant (R) genes on rice chromosome 12 including Pita, Pita2 and Ptr has been studies for decades. However, the relationship between these R genes has not been well established.
Results
In this study, we compared the resistance spectra controlled by Pita2 and Pita by testing their monogenic lines (MLs) in four hotspots found in the Philippines and Burundi from 2014 to 2018. The reaction patterns were distinct in two countries and that Pita2-mediated field resistance was relatively prevalent. Pathogenicity tests using 328 single-spore isolates in greenhouse further verified that IRBLta2-Re for Pita2 conferred a relatively broader spectrum resistance than those of Pita. Rough and fine mapping of Pita2 were conducted using F2 and F3 populations derived from IRBLta2-Re [CO] and CO 39 consisting of 4344 progeny to delimit Pita2 in a genomic interval flanked by two markers 12 g18530 and 12 g18920 proximal to the centromere of chromosome 12. Alignment of the markers to the genomic sequence of IR64, which harbors Pita2 verified by genetic analysis, approximately delimited the candidate gene(s) within 313-kb genomic fragment. The two Pita2 suppressive mutants that contain mutations within Pita2 were verified and identified. Comparative sequence analysis in these two mutants further identified that each individual allele contains a single nucleotide substitution at a different position resulting in nonsense and missense mutations in the protein product of LOC_Os12g18729. On the contrary, no sequence mutation was detected in other candidate genes, indicating that mutations in LOC_Os12g18729 were responsible for the loss of function of Pita2. Pita2 encodes a novel R protein unique from Pita, which is exactly identical to the previously cloned Ptr. Moreover, based on the resistance gene analysis of rice varieties and mutants containing Pita, it was found that Pita2 rather than Pita was responsible for the specificity to some differential isolates with AvrPita. The diagnosis and survey of Pita2 in IRRI released varieties showed relatively low frequency, implying a high value of its application for breeding resistant varieties against rice blast via marker assisted selection.
Conclusion
Our study clarified the relationship between Pita, Pita2 and Ptr. Pita2 is identical to Ptr and distinct from Pita in both sequence and chromosomal location although Pita2 and Pita are genetically linked to each other. The loss of function of Pita2 but not Pita eliminate the specificity to some AvrPita containing isolates, however, the mechanism underlying the recognition between Pita2/Pita and AvrPita remains elusive.
Similar content being viewed by others
Background
Plants have evolved two-branched of innate immune response to protect themselves against pathogens, including fungi, bacteria, viruses, and insects. The first branch recognizes pathogen-associated molecular pattern (PAMP) and triggers PAMP-triggered immunity (PTI), which generates the accumulation of reactive oxygen species (ROS) and the deposition of phenolic compounds (Jones and Dangl 2006). However, pathogens deliver effectors to suppress PTI (Bent and Mackey 2007). Resistant (R) genes in plant can recognize some of the effectors, referred to as avirulence (Avr) genes, and trigger the second branch of immune response called effector-triggered immunity (ETI), which can induce hypersensitive response (HR) to inhibit pathogen proliferation (Chisholm et al. 2006). Understanding the interaction between plants and pathogens can help to formulate better strategies to protect plants from diseases.
Rice blast caused by the fungal pathogen Magnaporthe oryzae destroys rice crops worldwide (Pennisi 2010). Incorporation of R genes into rice cultivars by marker assisted selection (MAS) presents an effective and economical way to control rice blast (Wang and Valent 2017). The bottleneck of this approach is that the R gene deployed in the varieties can be overcome in 2-4 years due to high level of variability in Avr genes of the pathogen (Skamnioti and Gurr 2009; Xiao et al. 2017). Therefore, continuous exploration of new R genes or alleles and utilization of effective ones in rice resistance breeding program are of great importance in controlling rice blast. As of now, more than 100 R genes have been mapped, 35 of which have been cloned (Wang et al. 2017). Except for Pid2, pi21 and Ptr, all of cloned R genes encode proteins which contain nucleotide binding site-leucine-rich repeat (NBS-LRR) domain (Skamnioti and Gurr 2009; Liu et al. 2010; Wang et al. 2017). Pi-d2 encodes a receptor-like kinase protein with a B-lectin and an intracellular serine-threonine kinase domain (Chen et al. 2006). pi21 encodes a protein that contains a putative heavy metal-binding domain (Fukuoka et al. 2009) while Ptr encodes a protein with Armadillo repeats domain (Zhao et al. 2018).
Rice blast R genes tend to be clustered, specifically on chromosome 6, 11 and 12 in rice (Liu et al. 2010). The cluster of R genes linked to the centromere on chromosome 12 includes Pita, Pita2 and at least eight other genes (Inukai et al. 1994; Yu et al. 1996; Li et al. 2008; Zheng et al. 2008; Koide et al. 2011; Lei et al. 2013; Dong et al. 2017). Pita2 was first reported in the variety Pi No4 and mapped on the centromeric region on chromosome 12 (Kiyosawa 1967; Rybka et al. 1997). Some reports suggest that Pita2 conferred a broader resistance spectrum than Pita (Kiyosawa 1967, 1971; Rybka et al. 1997). Pita2 was found resistant to all Pita avirulent isolates and some Pita virulent islolates (Kiyosawa 1967, 1971; Rybka et al. 1997). Furthermore, the resistance frequencies of Pita2 and Pita differential varieties (DVs) to 119 isolates in the Philippines are 84% and 25.2%, respectively (Yanoria et al. 2008). The relationship between Pita and Pita2 was reported to be closely linked or allelic (Rybka et al. 1997). Bryan et al. (2000) cloned Pita and proved that Pita2 specificity is a combination of Pita and another linked R gene (Bryan et al. 2000). However, it is complicated to clone Pita2 because all varieties containing Pita2 such as Reiho, Katy and IR64, were found to contain Pita (Lee et al. 2009; Zhao et al. 2018). The use of monogenic lines (MLs) and near-isogenic lines (NILs) of Pita2 makes it easier to map and clone this gene. Tsunematsu et al. (2000) developed the first set of 29 MLs for 24 R genes. Among them, IRBLta2-Re and IRBLta-CT2 have been used in many studies for Pita2 and Pita respectively (Lei et al. 2013; Mutiga et al. 2017). A series of NILs were developed under International Rice Research Institute (IRRI)-Japan Collaborative Research Project (Kobayashi et al. 2007; Yanoria et al. 2011). Of these NILs, IRBLta2-Re [CO] was one of the Pita2 NILs at BC6F21 derived from Reiho and CO39 (Yanoria et al. 2011). On the other hand, 20 isolates that can differentiate various R genes in MLs were selected from 119 isolates of the Philippines (Yanoria et al. 2008). Among them, V86010 is avirulent to Pita2 ML and can differentiate Pita and Pita2 (Yanoria et al. 2008). AvrPita was cloned and proved to trigger resistance in Pita2 rice as well as in Pita rice (Bryan et al. 2000; Orbach et al. 2000). Zhao et al. (2018) cloned Ptr which is required for Pita resistance and suggested that Ptr and Pita2 are indistinguishable (Zhao et al. 2018). The mapping and cloning of R genes in Pita cluster has made significant progress. However, the precise relationship between Pita, Pita2 and Ptr as well as the mechanism underlying the interaction between AvrPita and Pita, Pita2 and Ptr require further investigation. In this study, we aimed to clarify the relationship between Pita, Pita2 and Ptr. Cloning and uncovering the relationship of multiple genes in one cluster will be of great significance in elucidating the molecular interactions of these R genes, which will in turn guide the efforts of marker-assisted rice breeding for resistance.
Results
Distinct Reaction Patterns of Pita2 against Rice Blast Populations in the Philippines and Burundi
To assess the resistance spectrum of Pita2, IRBLta2-Re was used to monitor its performance in four disease hotspots found in the Philippines (Bohol) and Burundi (other three sites, Table 1). IRBLta2-Re showed stable resistance in Bohol from 2016 to 2018 (Table 1) and in all sites in Burundi from 2014 to 2016. However, IRBLta2-Re became susceptible in all sites in Burundi in both 2017 and 2018, indicating that Pita2 could have lost its effectiveness (Table 1). The distinct reaction patterns of Pita2 against rice blast populations in the Philippines and Burundi could result from different dynamics of pathogen populations in these two countries. Compared to the field performance of Pita2, Pita, which was represented by two MLs (IRBLta-Zh and IRBLta-CT2), showed susceptibility in all sites during all years monitored, except in Bohol in 2016 (Table 1).
Spraying inoculation study using single spore isolates showed that compared to other MLs, IRBLta2-Re showed relatively high resistance frequencies (RFs) ranging from 64% to 75% against 328 isolates collected in 2015 and 2017 (Table 2). In contrast, IRBLta-CT2 showed low RFs ranging from 32% to 35% (Table 2). The consistencies in the results obtained from the field studies with those from the greenhouse assessment using single spore isolates led us to conclude that the IRBLta2-Re controls a broader spectrum resistance than IRBLta-CT2 against rice blast isolates in the Philippines.
Genetic and Physical Delimitation of Pita2 Proximal to the Centromere of Chromosome 12
To genetically map Pita2, an F2 population derived from the cross between IRBLta2-Re [CO] and CO39 was generated. Of the 2151 F2 progeny inoculated with the Pita2-avirulent isolate V86010, 1607 and 544 were resistant and susceptible, respectively. The segregation ratio of resistance and susceptibility was found to fit a 3:1 in the χ2 test of a goodness-of-fit (χ2 = 0.096, P = 0.75), indicating that the resistance of IRBLta2-Re [CO], i.e. Pita2 against V86010, is controlled by a single locus/gene.
Two steps of mapping were employed to genetically delimit the location of Pita2. In the first step, 544 susceptible and 1001 resistant progeny from the F2 population mentioned above were used for the genetic analysis. Eleven recombinants, including five resistant and six susceptible progenies were identified between two SSR markers of RM27920 and RM1337 (Additional file 1: Table S1). The phenotype of the F3 descendant population derived from five resistant F2 progeny all showed segregation in resistance and susceptibility (Additional file 1: Table S1). Based on the phenotype and genotype analyses, eight and three recombinants were assigned at RM27920 and RM1337, respectively, from Pita2 (Fig. 1a and Additional file 1: Table S1). Six SNP markers between RM27920 and RM1337 were then developed and used for further screening of recombinants out of these 11 progeny, which helped to identify five and three recombinants at 12 g18120 and RM1337, respectively (Fig. 1a and Additional file 1: Table S1). No recombinants were identified at three markers within the genomic interval, indicating that Pita2 was flanked by markers of 12 g18120 and RM1337 (Fig. 1a). In the second step, the derived segregating F2 progeny resistant to V86010 was advanced to F3. A total of 2799 lines were screened for the recombinants between two CAPS markers, 12g18120F3/R4- NheI and 12g19304F2/R2- ClaI. Six recombinants were identified between 12 g18120 and 12 g19304 (Additional file 2: Table S2). After phenotyping the F4 descendant population derived from each recombinant F3 progeny, five and one recombinants were identified at 12 g18120 and 12 g19034, respectively from Pita2, indicating that Pita2 was delimited between these two markers (Fig. 1b and Additional file 2: Table S2). Six additional SNP markers were developed and used for the linkage analysis, which enabled the identification of four and one recombinants at 12 g18530 and 12 g18920, respectively, from Pita2 (Fig. 1b and Additional file 2: Table S2). No recombinants between 12 g18650 and Pita2 were identified, verifying the delimitation of Pita2 in this genomic interval (Fig. 1b and Additional file 2: Table S2). By aligning the two flanking markers (12 g18530 and 12 g18920) along the genomic sequence of Nipponbare, the delimited Pita2-containing genome interval was determined to be approximately 270 kb in length (position: 10,712,341 to 10,982,783 bp, http://rice.plantbiology.msu.edu) on chromosome 12. It is worth noting that the centromere of chromosome 12 was previously found residing in the BAC clone of OsJNBa0088J04, which spanned the genome interval at the positions from 11,871,508 to 12,007, 390 bp and was approximately 1 Mb apart from the putative Pita2 region.
Mapping and identification of Pita2.aPita2 was mapped between 12 g18120 and RM1337 on chromosome 12. bPita2 was fine mapped to 270 kb flanked by 12 g18530 and 12 g18920. The molecular markers and number of recombinants were listed above and underneath the thick line, respectively. The population size used for mapping is listed on the right. c Nine candidate genes of Pita2 were identified in the genomic region of IR64 delimited by markers of 12 g18530 and 12 g18920. The transcriptional direction was indicated by arrows. d Diagram of Pita2. Two mutants of IR64 were indicated by triangle. The position of amino acid residue corresponding to the single nucleotide mutation was indicated. Exons were showed in filled boxes. e Reaction pattern of different varieties to 9475-1-3. The figures was not drawn in scale
An F2 population derived from the cross between IR64 and CO39 was generated to validate whether the location of Pita2 in IR64 is located in the same genomic interval since IR64 was reported to harbor Pita2 and its genomic sequence is available. A total of 722 F2 progeny were inoculated with the Pita2-avirulent isolate 9475-1-3 and separated into 561 and 161 resistant and susceptible progeny, respectively. The ratio of the number of resistant versus susceptible progeny fitted perfectly 3:1 (χ2 = 2.81, P = 0.094), indicating that, most likely, there is only a single pair of R/Avr involved in the resistance of IR64 against 9475-1-3. Moreover, a CAPS marker (18729F3/R4-PvuII), which was derived from the gene model 12 g18729 and located between markers of 12 g18530 and 12 g18920, was developed and used for the genetic analysis (Fig. 2). All the 126 susceptible F2 progeny showed CO39 pattern, suggesting that Pita2 in IR64 was likely co-localized within the same genomic interval as identified in IRBLta2-Re [CO] (Fig. 2). The allelic region in IR64 was then retrieved from the IR64 genomic sequence (Schatz et al. 2014), which is approximately 313 kb in length, to further identify the candidate genes of Pita2.
Linkage analysis of R gene in IR64 to 9475-1-3 with Pita2 diagnosed CAPS marker (18729F3/R4- PvuII). F2 progeny derived from a cross between IR64 and CO39 were inoculated with 9475-1-3 and 126 susceptible lines were selected for linkage analysis. C1: IR64. C2: CO39. The CAPS marker has PvuII cleavage site on CO39 sequence. The product amplified by 18729F3/R4 from CO39 is 825 bp in size. After. PvuII cleavage, the products are 416 bp and 409 bp in size
Identification of Candidate Genes of Pita2 in IR64
We employed the program of Fgenesh (http://www.softberry.com) for gene prediction in the delimited region in IR64. A total of 43 gene models were identified, 17 of which corresponded to typical repetitive sequences (Additional file 3: Table S3). Moreover, other five gene models encoded small proteins with less than 100 amino acids in length. These two groups of gene models were not included in the further analysis. Except for two flanking genes, the sequences of the remaining 21 unique gene models were then compared to their alleles in the susceptible rice varieties, first with Nipponbare. Many of the 21 genes have their alleles with annotated gene models in Nipponbare. However, fours gene models, i.e., IR64–6, − 7, − 8, and − 16, were not predicted in Nipponbare. Further sequence analysis revealed that, except for IR64–6, the other three gene models were truly present in the genome sequence of Nipponbare albeit not predicted mainly due to frame shift sequence insertion/deletion. Sequence comparison between the gene models in IR64 and Nipponbare identified that the four gene models, IR64–28, − 34, − 36, − 38 were identical in protein sequence to LOC_Os12g18820, LOC_Os12g18860, LOC_Os12g18880, and LOC_Os12g18900 (http://rice.plantbiology.msu.edu/), respectively. Hence, these four gene models were not further considered as Pita2 candidates. To further shortlist the candidate genes, we compared the remaining 11 gene models to the genome sequence of another susceptible variety CO39 derived from the second generation of sequence assembly (GenBank accession number: SWLY00000000), which enabled the identification of other three gene models, namely, IR64–10, − 11, and − 26, to their respective alleles in CO39. Moreover, the three gene models IR64–4, − 9, and − 17 did not have sequence polymorphisms unique to their respective alleles in both Nipponbare and CO39, suggesting that these sequence polymorphisms could not be associated with the functional differences. Taken together, two rounds of sequence comparisons shortlisted nine gene models as candidates of Pita2 (Fig. 1c and Additional file 3: Table S3).
Identification of LOC_Os12g18729 as Pita2 by Characterization of Loss of Function Mutants Derived from EMS-Treated IR64 Mutant Population
Over 12,000 M2 individual plants were generated from EMS treated M1 plants of IR64 and screened to select susceptible mutants against 9475-1-3, the same isolate used for the genetic analysis of Pita2 in IR64. Two M2 lines, namely, #337 and #6599 that showed, respectively, 29:2 and 35:4 segregation ratio of resistance versus susceptibility in M2 progeny were identified. The susceptible progeny were then recovered and used for further characterization. First, genetic validation of these two mutants was conducted to determine whether the mutation occurred in the Pita2 gene itself or by its signaling components. These two mutants were crossed with CO39 to generate its respective F1 populations. Fifty-seven #337-derived and thirty-five #6599-derived F1 progeny were susceptible to 9475-1-3, indicating that CO39 could not complement the resistance in the F1 progeny. It can be speculated that the mutations in the Pita2 gene itself rather than in the putative signaling components in #337 and #6599 led to the loss of resistance of IR64. Secondly, a set of IRBLta2-Re avirulent isolates were selected to verify their reactions to both mutants. As shown in Tables 3, 13 out of 22 isolates reverted to virulence when inoculated with both #337 and #6599, indicating that IR64 triggered resistance against these isolates through a single R/Avr pair, i.e., Pita2/AvrPita2. The disruption of Pita2 resulted in the loss of function to those isolates. In contrast, additional R/Avr gene pairs to Pita2/AvrPita2 existed in the interaction between IR64 and other nine isolates (IK81–25, M101–1–2-9-1, B90036, B90002, B90165, B90324, V850196, V86010 and C9240–4). Lastly, sequencing showed that the gene model IR64–18, i.e., LOC_Os12g18729, contained sequence mutations in different positions in #337 and #6599 compared to the one in the wild type IR64. The point mutation from guanine (G) to thymine (T) in LOC_Os12g18729 in #337 resulted in the amino acid change from aspartic acid (Asp) to tyrosine (Tyr) at 124th position. In #6599, the point mutation from cytosine (C) to thymine (T) resulted in the generation of a premature stop codon from glutamine (Gln) in LOC_Os12g18729 at the 213rd amino acid position (Fig. 1d and e). On the contrary, no sequence mutations were identified in the other eight candidate genes. Therefore, we postulated that the different mutations of LOC_Os12g18729 caused the loss of function of Pita2 in #337 and #6599, and that LOC_Os12g18729 was most likely responsible for the function of Pita2.
Pita2 Encodes a Unique R Protein and Is Identical to Ptr
The predicted coding sequence of Pita2 in IR64 is 2718 bp in length and composed of four exons (Fig. 1d), which were verified by sequencing the transcript obtained from the 3-week-old seedling (data not shown). The derived protein product contained 905 amino acid residues, and predicted to contain three strong transmembrane helices located at positions 85–106, 165–185, and 214–232 amino acids (https://embnet.vital-it.ch/cgi-bin/TMPRED_form_parser). However, the subcellular localization of Pita2 was predicted inconsistently by different programs either in the cytoplasm (https://www.predictprotein.org/visual_results?req_id=686316ic) or extracellularly (http://www.softberry.com/cgi-bin/programs/proloc/protcomppl.pl). It is worth noting that Pita2 was identical to Ptr as described previously, which was found to contain an Armadillo (ARM) repeat domain (Zhao et al. 2018). No significant sequence similarity was found between Pita and Pita2. The expression of Pita2 in IRBL28, CN16, and IR64 was investigated and found that Pita2 was expressed constitutively at different time points after infection of rice blast (Additional file 4: Fig. S1). The constitutive expression of Pita2 was consistent with the finding described previously (Zhao et al. 2018).
Analysis of Pita2 Haplotypes in IRBLs Containing Pi19 or its Tentative Alleles
A comprehensive analysis of haplotype diversity of Ptr was conducted previously (Zhao et al. 2018). In this study, we investigated the sequence variation of Pita2 haplotypes in 4 monogenic lines (IRBLkp-K60, IRBLzt-T, IRBLta-K1, IRBL19-A) which were characterized to each contain Pi19 or its functional allele previously (Selisana et al., 2017) and two susceptible rice varieties (LTH and CO39). The Pita2 haplotypes in all these 6 rice varieties were found to belong to the haplotype group containing a 12-bp insertion (Table 4), which was characterized to be associated with the pathogen recognition specificity as described previously (Zhao et al. 2018). In addition to this 12-bp insertion, they contained a varied number of residues different from Pita2 in Katy or IR64 (Table 4). Sequence comparison further revealed that some rice varieties contained identical Pita2 haplotypes. For example, Nipponbare and IRBLzt-T contained an identical Pita2 haplotype whereas IRBL19-A and IRBLkp-K60 contained another identical Pita2 haplotype (Table 4 and Additional file 5: Fig. S2). It is worth noting that the Pita2 haplotype in CO39, which was more closely related to Pita2 in IR64 or Katy (Additional file 5: Fig. S2), was found to be responsible for the resistance of CO39 to some avirulent isolates based on the functional analysis of CRISPR line (Gui et al., unpublished), implying that the Pita2 haplotype in CO39 is a novel functional allele of Pita2. It is elusive whether other Pita2 haplotypes identified in four monogenic lines containing Pi19 or its alleles are responsible for the resistance of their respective varieties.
Pita2 rather than Pita Conferred Resistance to some AvrPita-Containing Isolates
The identification of Pita2 revealed that Pita2 (corresponding to LOC_Os12g18729) and Pita (corresponding to LOC_Os12g18360) were encoded by different genes and separated from each other by approximately 210 kb in the genome sequence of Nipponbare. The recombinant 8-2B3, which was heterozygous at the Pita2 locus but homozygous with the susceptible allele at the Pita locus, verified that the function of Pita2 was independent from Pita (Additional file 2: Table S2). To clarify the contribution of resistance spectrum conferred by Pita and Pita2, two MLs and one cultivar (IRBLta-CT2, IRBLta2-Re, IR64) were used for resistance spectrum analysis. The presence of Pita and Pita2 were verified by PCR amplification and subsequent amplicon sequencing, which revealed that both IR64 and IRBLta2-Re contained both Pita and Pita2 as described previously (Lee et al. 2009; Zhao et al. 2018). However, IRBLta-CT2 contained only Pita as reported previously (Lee et al. 2009). A single T base-pair deletion corresponding to the position of 554 in the coding sequence of Pita2 in IR64 was identified in IRBLta-CT2, leading to a frame shift in the coding sequence and a disrupted allele of Pita2. A set of seven isolates containing AvrPita was used for pathogenicity test using these rice varieties and the susceptible check variety, LTH (Fig. 3). Table 3 shows that both IR64 and IRBLta2-Re were resistant to all isolates. Intriguingly, IRBLta-CT2 was susceptible to all isolates except IK81–25 (Fig. 3), suggesting that Pita in IRBLta-CT2 did not function as it did in IRBLta2-Re and IR64. It is reasonable to assume that additional R gene(s) could contribute to the resistance of IRBLta-CT2 to IK81–25. Moreover, it is worth noting that two Pita2 disrupted mutants (#337 and #6599) were susceptible to four of these AvrPita isolates, except IK81–25 (other three isolates were not included in the analysis), indicating that disruption of Pita2 resulted in the loss of resistance to those isolates (Table 3). However, it is still elusive whether this change from resistance to susceptibility of the mutants to the AvrPita isolates is due to the recognition between Pita2 and AvrPita or another unknown avirulence effector.
The Frequency of Pita2 in Modern IRRI Released Varieties
To investigate the frequency of Pita2 in improved varieties, we used the InDel marker for Ptr, Z12F/R (Zhao et al. 2018) in the diagnosis of a set of 46 IRRI varieties. There were 14 out of 46 selected IRRI varieties resolved a PCR amplicon similar to that of IR64 (Table 5 and Additional file 6: Fig. S3). On the contrary, 32 varieties resolved an amplicon similar to that of CO39, which was 12 bp larger than that of IR64 by referring to their sequence differences of alleles of Pita2. A set of seven differential isolates, including five avirulent and two virulent isolates to Pita2, was used to evaluate the resistance spectrum of these 46 varieties. The rice lines that resolved a smaller amplicon were resistant to all five Pita2-avirulent isolates (Table 5 and Additional file 6: Fig. S3). However, at least one of the five isolates was virulent to the rice lines resolving a larger amplicon. We speculate that the haplotype of Pita2 present in the 34 rice varieties that resolved a larger amplicon did not confer the function of Pita2 as identified in IR64. In this regard, the frequency of Pita2 in IRRI released varieties was approximately 30%. The sequence corresponding to the 4th exon of Pita2 where the InDel resided was further amplified using the primer pair 18729F3/R4 and subsequently determined. The result identified three different haplotypes in the 34 rice varieties. The first haplotype was found in two varieties (NSIC Rc 220SR and NSIC Rc 304SR) identical to that of CO39. The second haplotype was found in six varieties (NSICRc300, NSICRc340, NSICRc192, NSICRc280, NSICRc288 and NSICRc342) that contained a fragment deletion outside the InDel. The last one was found in 26 rice varieties that contained a single nucleotide change (from “G” to “C”) when compared to that of CO39. Intriguingly, this nucleotide change depleted the cleavage site of PvuII. Thus, CAPS marker (18729F3/R4-PvuII) was developed to differentiate these haplotypes (Table 5 and Additional file 6: Fig. S3) and can be used for the introgression of Pita2 via marker assisted selection.
Discussion
Pita2 encodes a Sequence and Genomic Location Unrelated R Protein of Pita
Pita2 was firstly reported in the variety Pi No4, which was allelic to Pita in another variety, Pi No1, and mapped to the centromeric region on chromosome 12 (Kiyosawa 1967; Rybka et al. 1997). Several other R genes such as Pi-4a(t), Pi-4b(t), Pi-6(t), and Pi-12(t), Pi57(t), Pi19(t), Pi61(t), Pi20(t) were also identified to be allelic or linked to Pita and Pita2 (Inukai et al. 1994; Yu et al. 1996; Li et al. 2008; Zheng et al. 2008; Koide et al. 2011; Lei et al. 2013; Dong et al. 2017). The cloning of Pita revealed that it encoded an NBS-LRR type R protein, which made a significant progress in the dissection of this genetic complex harboring multiple R genes (Bryan et al. 2000). Zhao et al. (2018) recently reported the cloning of a new broad-spectrum R gene Ptr, which encoded a novel R protein containing an ARM repeat domain. Ptr was found to be required for the function of Pita and its disruption depleted the function to AvrPita-containing strains (Zhao et al. 2018). However, the relationship between Pita and Pita2 as well as other R genes remains elusive. In this study, we mainly utilized genetic approach to differentiate the Pita2 gene in the Pita2 containing varieties, IRBLta2-Re [CO], IR64 and IRBLta2-Re. The use of a large genetic population enabled the delimitation of Pita2 into a small genomic interval. More importantly, the recombinants between Pita2 and Pita which are identified in the population broke the association of Pita2 from Pita, allowing the clarification of the relationship between them. For example, the identification of the recombinant 8-2B3 verified that Pita2 was independent from Pita (Additional file 2: Table S2). Moreover, the generation and characterization of two Pita2 independent suppressor mutants provided convincing evidence that the single gene/locus conferring resistance in IR64 against the isolates was controlled by Pita2. The set of isolates avirulent to the IRBLta2-Re which was often used for the diagnosis of the effectiveness of Pita2 in the field and green house gained virulence to two Pita2 suppressor mutants, further verifying that resistance of IR64 against these diagnostic isolates was attributable to Pita2 rather than on other R genes in IR64 (Table 3). The molecular characterization of Pita2 confirmed that it was distinct from Pita based on the sequence and genomic location, although they were tightly linked in the approximately 200 kb genomic interval, clarifying the genetic relationship between Pita and Pita2. The allelism test among Pi19 in IRBL19-A, Pi20 in IRBL20-IR24, and Pita2 in IRBLta2-Re revealed that Pi19, Pi20, and Pita2 were most likely allelic to each other (unpublished), implicating that Pi19 and Pi20 could be alleles of Pita2. It was found that Ptr being verified as Pita2 in this study contained abundant variations in diverse rice germplasm (Zhao et al. 2018). The molecular characterization of Pita2 paved a way in functional characterization of these haplotypes using sequence-based approaches, e.g., CRISPR and gene complementation tests.
How Pita2 and Pita Contribute to the Specificity to Isolates Containing AvrPita Is Still Elusive
Based on pathogenicity tests in both field and greenhouse, it is demonstrated that IRBLta2-Re showed higher resistance frequency than IRBLta-CT2, indicating that Pita2 conferred a broader spectrum resistance than Pita. Previous studies indicated that Pita resistance was also found in Pita2 varieties, suggesting Pita2 resistance specificity could be a combination of Pita and at least another R gene (Bryan et al. 2000). Intriguingly, the loss of function of Ptr compromised the resistance of both Pita and Pita2, suggesting that Ptr was required for the function of Pita (Zhao et al. 2018). In this study, it is demonstrated that the isolates with AvrPita gained virulence to Pita2 suppressor IR64 mutants and IRBLta-CT2 in which Pita2 was truncated due to a single nucleotide deletion in the coding region (Table 3 and Fig. 3), indicating that Pita2 was required for the function of Pita. The verification of Ptr being Pita2 explained the findings of this study. In this regard, the resistance observed in IRBLta-CT2 could be not attributable to Pita, which may result in the complexity in the comparison of resistance spectrum between Pita and Pita2. However, it is doubtless that Pita2 can also recognize non-AvrPita avirulence effectors in rice blast pathogen. For example, AvrPita was not identified in both V86010 and 9475-1-3 that were used in the genetic analysis of Pita2 in this study. The hypothesis that Pita2 is required for the function of Pita needs to be further verified. The knockout of Pita in rice varieties containing both Pita and Pita2 can be helpful to finish the remaining pieces in this puzzle. It is also important to use transformed isolates with and without AvrPita to investigate their pathogenicity to rice lines with and without Pita2 and Pita.
Molecular Cloning of Different R Genes Clustered in the Same Genomic Complex Is Fundamentally Important for Precise Identification and Utilization in Resistance Breeding Program
Over 100 rice blast R genes were genetic mapped and designated in different rice varieties (Zhao et al. 2018). Due to the complexity of R genes and their clusters, less than 30 R genes were characterized at the molecular level (Su et al. 2015; Zhao et al. 2018). Due to the use of different set of isolates usually collected locally, the comparison of resistance spectrum conferred by different R genes becomes an impossible task. Thus, new R genes identified in unique donors are often designated to a new name using either a number following the latest one or the name of the origin as the suffix after the abbreviation of Pi. Moreover, many R genes with different designation reported by different groups were actually identical to each other. For example, in our previous study, we found that the broad-spectrum resistance observed in JaoHom Nin, one of the widely used donor for blast resistance, was controlled by two known R genes, Pi7 and Pish (Chaipanya et al. 2017). It was also found that Pigm was actually identical in sequence to Pi50 cloned earlier (Su et al. 2015; Deng et al. 2017). A comprehensive screening of Pi2/9 alleles identified at least four rice varieties contained Pi2-A15 identical to Pi50/Pigm based on sequence comparison (Xiao et al. 2017). It is therefore reasonable to assume that new R genes with different designation could be allelic or even identical to the previously characterized R genes. Thus, it is necessary to have a more standard nomenclature system to avoid the confusion particularly to rice breeders in selecting the correct genes in their breeding program. The sequence verification of known R genes linked to the newly identified R genes must be a prerequisite before a new gene designation is assigned.
In this study, the cloning of Pita2 clarified its relationship to Pita and Ptr. Comprehensive sequence analysis of their haplotypes in the donor varieties containing Pita/Pita2 linked R genes previously reported by different groups has effectively clarified the relationship of these R genes with Pita and Pita2. In this study, we identified three Pita2 haplotypes in 46 IRRI varieties containing sequence differences from Pita2. The varieties containing these Pita2 haplotypes were susceptible to Pita2-avirulent isolates. Sequences of these haplotypes were referred in the development of InDel and CAPS markers that can be used for the introgression of Pita2 in these susceptible rice varieties.
Conclusion
In this study, Pita and Pita2 resistance spectra were assessed in the field and in the greenhouse. Pita2 conferred a broader resistance spectrum than Pita. Pita2 was mapped approximately 313 kb genomic interval flanked by 12 g18530 and 12 g18920 on the short arm of chromosome 12 based on the equivalent sequence of IR64. LOC_Os12g18729 was verified as a single candidate of Pita2 by characterizing suppressive mutants. The IRRI newly released varieties were assessed by Pita2 diagnostic markers, which implicate that Pita2 has a high value in future rice breeding. A novel Pita2 allele was found in many IRRI varieties that have a single nucleotide change (from “G” to “C”) compared to the allele in CO39. A CAPS marker (18729F3/R4-PvuII) was developed that can distinguish different Pita2 haplotypes. Taken together, our study clarifies the relationship between Pita, Pita2 and Ptr. Moreover, the developed markers will be useful for the improvement of future blast resistance rice breeding programs.
Methods and Materials
Plant Materials and Isolates
Plant materials used in this study were listed in Additional file 7: Table S4. Twenty-three IRBLs harboring different resistant R genes in the background of LTH (Tsunematsu et al. 2000; Yanoria et al. 2010) and IRBLta2-Re [CO] harboring Pita2 in the background of CO39 (Yanoria et al. 2011) were maintained at IRRI (https://www.irri.org). IR64 and 46 IRRI new released varieties were obtained from International rice genebank collection (IRGC) of IRRI. Three hundred and fifty-five M. oryzae isolates were collected in the different years and maintained at rice blast isolate collection at IRRI (Additional file 8: Table S5).
Disease Evaluation in the Greenhouse and Field
The disease evaluation in the greenhouse followed Yanoria et al. (2008). Briefly, isolates were cultured in prune agar medium (three pieces prunes, 5 g alpha-lactose monohydrate, 1 g yeast extract, 21 g gulaman bar and 1 L distilled H2O) and spores were harvested using 0.02% (vol/vol) Tween 20 in distilled water. The final spore concentration for inoculation is 1 × 105 conidia/ml. Rice seedlings at the three-to four-leaf stage were used for blast inoculation. Disease assessment was done 6-7 days after inoculation following the method of Bonman (1986) wherein 0 = no evidence of infection; 1 = brown specks smaller than 0.5 mm in diameter, no sporulation; 2 = brown specks about 0.5-1 mm in diameter, no sporulation; 3 = roundish to elliptical lesions about 1–3 mm in diameter with gray center surrounded by brown margins, lesions capable of sporulation; 4 = typical spindle-shaped blast lesions capable of sporulation, 3 mm or longer with necrotic gray centers and water-soaked or reddish brown margins, little or no coalescence of lesions; 5 = lesions as in 4 but about half of one or two leaf blades killed by coalescence of lesions. Scores of 0–3 were considered resistant reactions, and scores of 4 and 5 were considered susceptible reactions.
For field evaluation, four hotspots were chosen for monitoring: Bohol, in the Philippines and Gisha, Cankuzo and Rugombo, in Burundi. Three replications were set for every hotspot wherein each replicate containing 50 lines. Disease evaluation was done 40 days after transplanting based on the Standard Evaluation System (SES) IRRI (2013): 0–1: resistant; 3: moderately resistant; 5: moderately susceptible; 7: susceptible; 9: very susceptible.
Designing Markers
Two SSR markers (RM27920 and RM1337) which were reported close to the centromere of chromosome 12 (Jia et al. 2012) and had polymorphism between IRBLta2-Re [CO] and CO39 were selected for rough mapping. For fine mapping Pita2, SNP and CAPS markers within delimited interval were designed. To make sure that the designed markers are specific on chromosomes, annotated expressed proteins in “rice functional genomic express database” (http://signal.salk.edu/cgi-bin/RiceGE5) were selected. Allelic region of these selected genes in IR64 and CO39 were compared and markers were designed based on the polymorphism between these two genomes. The SNP markers 12 g18120 and 12 g19304 were further developed into CAPS markers to screen more recombinants (Additional file 9: Table S6).
Sequence Analysis
The polymorphic sequences in recombinants were amplified by PCR using SNP markers (Additional file 9: Table S6). Coding sequence of nine Pita2 candidate genes in IR64, #337 and #6599 were amplified by PCR using primers (Additional file 9: Table S6). Sequencing was conducted by Biosune (http://www.biosune.com/) in China. BLASTN search program (https://blast.ncbi.nlm.nih.gov/Blast) and “sequencher 5.4.6” software (http://www.genecodes.com) were used for sequence alignment.
EMS Mutagenesis and Mutants Screening
Approximately 12,000 seeds of IR64 were pre-soaked in an opaque bucket containing four liters of distilled water. This was incubated at 28 °C for 24 h. After incubation, the water was drained and replaced with a freshly prepared 4-l 0.8% EMS solution. The bucket was gently mixed to ensure that the solution would deeply penetrate the seeds and was incubated for 16 h. The mutagen solution was drained after incubation and the treated seeds were washed three times with distilled water. The solution and all the washings were collected for inactivation. Treated seeds were sown in pre-fertilized soil and allowed to grow for 21 days. The seedlings were transplanted in the field and the normal breeder’s practice was implemented for the field maintenance. The seedlings were allowed to grow until maturity. Seeds from surviving plant were collected as M2 population of each mutant line. Segregation analysis was done on M2 of each mutant line using isolate 9475-1-3.
A High Throughput Protocol for Plant Genomic DNA Preparation and PCR
Rice seeds were pre-soaked for 48 h and sown in a 96-well PCR plate with cut bottom. Then, the PCR plate was kept in a plastic tray (60 cm × 38 cm × 12 cm) and plants were grown hydroponic solution (Shouichi et al. 1976). After 1 week, fresh leaves were cut into small pieces (1 mm wide) and place in another 96-well PCR plate with 40ul TPS buffer (100 mM Tris-HCl (PH8.0) 100 ml, 10 mM EDTA 20 ml, 1 M KCl 74.55 g, add water up to 1 L and sterilize it by autoclave). The mixture in PCR plate was then incubated at 95 °C for 5 min to extract DNA. LA Taq DNA polymerase (http://www.takara-bio.com/) was used following this system (10 x LA taq buffer 1ul, dNTP 1ul, DNA mixture 1.5ul, LA taq enzyme 0.1ul, forward primer 0.25ul, reverse primer 0.25ul, ddH2O 5.9ul). PCR was done following this PCR profile (95° 2 min, 98° 10s, 55° 30s, 68° 30s, 72° 10 min, 33 cycles).
Availability of Data and Materials
Not applicable.
Abbreviations
- ARM:
-
Armadillo
- Avr:
-
Avirulence
- DV:
-
Differential Varieties
- ETI:
-
Effector Triggered Immunity
- HR:
-
Hypersensitive Response
- LRR:
-
Leucine-Rich Repeat
- MAS:
-
Marker Assisted Selection
- ML:
-
Monogenic Line
- NBS:
-
Nucleotide Binding Site
- NIL:
-
Near Isogenic Line
- PAMP:
-
Pathogen Associated Molecular Pattern
- PCR:
-
Polymerase Chain Reaction
- PTI:
-
PAMP -Triggered Immunity
- R gene:
-
Resistance gene
- ROS:
-
Reactive Oxygen Species
References
Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45:399–436
Bonman JM (1986) Physiologic specialization of Pyricularia oryzae in the Philippines. Plant Dis 70:767–769
Bryan GT, Wu KS, Farrall L, Jia Y, Hershey HP, McAdams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12:2033
Chaipanya C, Yanoria MJT, Quime B, Longya A, Korinsak S, Korinsak S, Toojinda T, Vanavichit A, Jantasuriyarat C, Zhou B (2017) Dissection of broad-spectrum resistance of the Thai rice variety Jao Hom Nin conferred by two resistance genes against rice blast. Rice 10:18
Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, Ma B, Wang Y, Zhao X, Li S, Zhu L (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46:794–804
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124:803–814
Deng Y, Zhai K, Yang D, Zhu X, Liu J, Wang X, Qin P, Yang Y, Zhang G, Li Q, Zhang J, Wu S, Milazzo J, Mao B, Wang E, Xie H, Tharreau D, He Z (2017) Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355:962–965
Dong L, Liu S, Xu P, Deng W, Li X, Tharreau D, Li J, Zhou J, Wang Q, Tao D, Yang Q (2017) Fine mapping of Pi57(t) conferring broad spectrum resistance against Magnaporthe oryzae in introgression line IL-E1454 derived from Oryza longistaminata. PLoS One 12:1–11
Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M (2009) Loss of function of a proline containing protein confers durable disease resistance in rice. Science 325:998–1001
Inukai T, Nelson RJ, Zeigler RS, Sarkarung S, Mackill DJ, Bonman JM, Takamure I, Kinoshita T (1994) Allelism of blast resistance genes in near-isogenic lines of rice. Phytopathology 84:1278–1283
IRRI (2013) Standard evaluation system for Rice (SES). International Rice Research Institute, Manila, p18
Jia Y, Jia MH, Wang X, Liu G (2012) Indica and japonica crosses resulting in linkage block and recombination suppression on rice chromosome 12. PLoS One 7:e4306610
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kiyosawa S (1967) Inheritance of resistance of the rice variety PI no. 4 to blast. Jpn J Breed 17:165–172
Kiyosawa S (1971) Gene analysis of blast resistance in exotic varieties of rice. Jarq 6:8–15
Kobayashi N, Yanoria MJT, Tsunematsu H, Kato H, Imbe T, Fukuta Y (2007) Development of new sets of international standard differential varieties for blast resistance in rice (Oryza sativa L.). Crop Sci 41:31–37
Koide Y, Yanoria MJT, Dela PF, Fukuta Y, Kobayashi N (2011) Characterization of Rice blast isolates by the differential system and their application for mapping a resistance gene, Pi19(t). J Phytopathol 159:85–93
Lee S, Costanzo S, Jia Y, Olsen KM, Caicedo AL (2009) Evolutionary dynamics of the genomic region around the blast resistance gene pi-ta in AA genome oryza species. Genetics 183:1315–1325
Lei C, Hao K, Yang Y, Ma J, Wang S, Wang J, Cheng Z, Zhao S, Zhang X, Guo X, Wang C, Wan J (2013) Identification and fine mapping of two blast resistance genes in rice cultivar 93-11. Crop J 1:2–14
Li W, Lei C, Cheng Z, Jia Y, Huang D, Wang J, Wang J, Zhang X, Su N, Guo X, Zhai H, Wan J (2008) Identification of SSR markers for a broad-spectrum blast resistance gene Pi20(t) for marker-assisted breeding. Mol Breed 22:141–149
Liu J, Wang X, Mitchell T, Hu Y, Liu X, Dai L, Wang GL (2010) Recent progress and understanding of the molecular mechanisms of the rice- Magnaporthe oryzae interaction. Mol Plant Pathol 11:419–427
Mutiga SK, Rotich F, Ganeshan VD, Mwongera DT, Mgonja EM, Were VM, Harvey JW, Zhou B, Wasilwa L, Feng C, Ouédraogo I, Wang GL, Mitchell TK, Talbot NJ, Correll JC (2017) Assessment of the virulence spectrum and its association with genetic diversity in Magnaporthe oryzae populations from sub-Saharan Africa. Phytopathology 107:852–863
Orbach MJ, Farrall L, Sweigard JA, Chumley FG, Valent B (2000) A telomeric avirulence gene determines efficacy for the rice blast resistance gene pi-ta. Plant Cell 12:2019–2032
Pennisi E (2010) Armed and dangerous. Science 327:804–805
Rybka K, Miyamoto M, Ando I, Saito A, Kawasaki S (1997) High resolution mapping of the Indica -derived rice blast resistance genes II. Pi-ta2 and Pi-ta and a consideration of their origin. Mol Plant-Microbe Interact 10:517–524
Schatz MC, Maron LG, Stein JC, Wences AH, Gurtowski J, Biggers E, Lee H, Kramer M, Antoniou E, Ghiban E, Wright MH, Chia J, Ware D, McCouch SR, McCombie WR (2014) Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol 15:506
Selisana SM, Yanoria MJ, Quime B, Chaipanya C, Lu G, Opulencia R, Wang G-L, Mitchell T, Correll J, Talbot NJ, Leung H, Zhou B (2017) Avirulence Gene-Based Diagnosis Complements Existing Pathogen Surveillance Tools for Effective Deployment of Resistance Genes Against Rice Blast Disease. Phytopathology 107(6):711–720
Shouichi Y, Cock JH, Gomez K (1976) Routine procedure for growing rice plants in culture solution. In: laboratory manual for physiological studies of rice, 3rd edn. International Rice Research Institute, Manila, p 62
Skamnioti P, Gurr SJ (2009) Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol 27:141–150
Su J, Wang W, Han J, Chen S, Wang C, Zeng L, Feng A, Yang J, Zhou B, Zhu X (2015) Functional divergence of duplicated genes results in a novel blast resistance gene Pi50 at the Pi2/9 locus. Theor Appl Genet 128:2213–2225
Tsunematsu H, Yanoria MJT, Ebron LA, Hayashi N, Ando I, Kato H, Imbe T, Khush GS (2000) Development of monogenic lines of rice for blast resistance. Breed Sci 50:229–234
Wang BH, Ebbole DJ, Wang ZH (2017) The arms race between Magnaporthe oryzae and rice: diversity and interaction of Avr and R genes. J Integr Agric 16:2746–2760
Wang GL, Valent B (2017) Durable resistance to rice blast. Science 355:906–907
Xiao G, Borja FN, Mauleon R, Padilla J, Yanoria MJT, Yang J, Lu G, Dionisio-Sese M, Zhou B (2017) Identification of resistant germplasm containing novel resistance genes at or tightly linked to the Pi2/9 locus conferring broad-spectrum resistance against rice blast. Rice 10:37
Yanoria MJT, Imbe T, Kato H, Tsunematsu H, Ebron LA, Vera Cruz CM, Kobayashi N, Fukuta Y (2008) A set of standard differential blast isolates (Magnaporthe grisea (Hebert) Barr.) from the Philippines for rice (Oryza sativa L.) resistance. Jarq Jpn Agr Res Q 42:23–34
Yanoria MJT, Koide Y, Fukuta Y, Imbe T, Kato H, Tsunematsu H, Kobayashi N (2010) Development of near-isogenic lines of japonica-type rice variety Lijiangxintuanheigu as differentials for blast resistance. Breed Sci 60:629–638
Yanoria MJT, Koide Y, Fukuta Y, Imbe T, Tsunematsu H, Kato H, Ebron LA, Nguyen TMN, Kobayashi N (2011) A set of near-isogenic lines of Indica-type rice variety CO39 as differential varieties for blast resistance. Mol Breed 27:357–373
Yu ZH, Mackill DJ, Bonman JM, SR MC, Guiderdoni E, Notteghem J-L, Tanksley SD (1996) Molecular mapping of genes for resistance to rice blast (Pyricularia grisea Sacc.). Theor Appl Genet 93:859–863
Zhao H, Wang X, Jia Y, Minkenberg B, Wheatley M, Fan J, Jia MH, Famoso A, Edwards JD, Wamishe Y, Valent B, Wang GL, Yang Y (2018) The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat Commun 9:2039
Zheng KL, Zhuang JY, Lu J, Qian HR, Lin HX (2008) Identification of DNA markers tightly linked to blast resistance genes in rice. In: Rice Genetics III. World Scientific Publishing Company, pp 565–569
Acknowledgements
Not applicable.
Funding
This research was partly supported by “National Natural Science Foundation of China (31461143019)”, “International Scientific and Technological Cooperation and Exchange Project of Fujian Agriculture and Forestry University (KXGH17007)”, “China Scholarship Council/ International Rice Research Institute Joint Scholarships”, and “the Swiss Programme for Research on Global Issues for Development (IZ07Z0-160877)”.
Author information
Authors and Affiliations
Contributions
XM, BZ and GL conceived and designed the experiments. XM, GX, MJTY, PMS, JP, JB, JW and ML performed the experiments. XM, GX, MJTY, RO, GH, BW, GL and BZ analyzed the data. XM, MJTY, RO, GL and BZ wrote the manuscript. All authors read and approved the final manuscript.
Authors’ Information
Not applicable.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Genotype and phenotype of 11 recombinants in the first round of genetic analysis
Additional file 2: Table S2.
Genotype and phenotype of 6 recombinants in the second round of genetic analysis
Additional file 3: Table S3.
Candidate genes of Pita2
Additional file 4: Fig. S1.
Expression pattern of Pita2 haplotypes in different rice varieties.
Additional file 5: Fig. S2.
Phylogenetic tree analysis of the Pita2 haplotypes in different rice varieties.
Additional file 6: Fig. S3.
Forty-six IRRI new released varieties were diagnosed by two Pita2 markers.
Additional file 7: Table S4.
Plant materials
Additional file 8: Table S5.
Isolates
Additional file 9: Table S6.
Markers
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Meng, X., Xiao, G., Telebanco-Yanoria, M.J. et al. The broad-spectrum rice blast resistance (R) gene Pita2 encodes a novel R protein unique from Pita. Rice 13, 19 (2020). https://doi.org/10.1186/s12284-020-00377-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-020-00377-5