Abstract
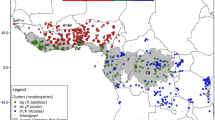
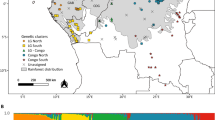
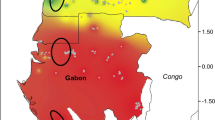
Species delimitation remains a crucial issue for widespread plants occurring across forest-savanna ecotone such as Lophira (Ochnaceae). Most taxonomists recognize two parapatric African tree species, widely distributed and morphologically similar but occurring in contrasted habitats: L. lanceolata in the Sudanian dry forests and savannahs and L. alata in the dense Guineo-Congolian forests. Both species co-occur along a ca. 3000 km long forest-savanna mosaic belt, constituting ideal models for investigating hybridization patterns and the impact of past glacial periods on the genetic structures in two types of ecosystems. We genotyped 10 nuclear microsatellites for 803 individuals sampled across the distribution range of Lophira. Both species exhibit similar levels of genetic diversity [He = 0.52 (L. alata); 0.44 (L. lanceolata)] and are well differentiated, consistent with taxonomic delimitation (FST = 0.36; RST = 0.49), refuting the hypothesis that they might constitute ecotypes rather than distinct species. Furthermore, L. alata displayed two deeply differentiated clusters (FST = 0.37; RST = 0.53) distributed in parapatry, one endemic to Western Gabon while another cluster extended over the remaining species range, suggests that L. alata is made of two cryptic species. We showed that rare hybrids occur in some contact zones between these three species, leaving a weak signal of introgression between L. lanceolata and the northern cluster of L. alata. At the intra-specific level, the latter species also show weak genetic structuring between Upper and Lower Guinea and the intensity did not differ strikingly between rainforest and savanna ecosystems. The discovery of a new species of Lophira with a narrow distribution in West Gabon where it is intensively exploited for its timber requires to evaluate its conservation status.



Similar content being viewed by others
References
Akoegninou A, van der Burg WJ, van der Maesen JG (2006) Flore analytique du Bénin. Backhuys Publishers, Leiden
Allal F, Sanou H, Millet L, Vaillant A, Camus-Kulandaivelu L, Logossa ZA et al (2011) Past climate changes explain the phylogeography of Vitellaria paradoxa over Africa. Heredity 107:601. https://doi.org/10.1038/hdy.2011.112
Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160:1217–1229
Anon, Chev (1954) Monographie de Azobé, Lophira procera A. Centre Technique Forestier Tropical, Nogent-sur-Marne
Aubreville A (1959) La flore forestière de la Côte d’Ivoire, 2nd ed. Centre Technique Forestier Tropical, Nogent-sur-Marne, p 958
Bamps P (1970) Répartition géographique du genre Lophira Banks ex Gaertn. (Ochnaceae). Bull Jard Bot Belg 40:291–294
Biwole A, Bourland N, Daïnou K, Doucet J-L (2012) Définition du profil écologique de l’azobé, Lophira alata, une espèce ligneuse africaine de grande importance: synthèse bibliographique et perspectives pour des recherches futures. Biotechnol Agron Soc Environ 16:217–228
Born C, Alvarez N, McKey D, Ossari S, Wickings EJ, Hossaert-McKey M et al (2011) Insights into the biogeographical history of the Lower Guinea Forest Domain: evidence for the role of refugia in the intraspecific differentiation of Aucoumea klaineana. Mol Ecol 20:131–142
Channan S, Collins K, Emanuel WR (2014) Global mosaics of the standard MODIS land cover type data. University of Maryland and the Pacific Northwest National Laboratory, College Park
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Chybicki IJ, Burczyk J (2009) Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 100:106–113
Daïnou K, Blanc-Jolivet C, Degen B, Kimani P, Ndiade-Bourobou D, Donkpedan ASL et al (2016) Revealing hidden species diversity in closely related species using nuclear SNPs, SSRs and DNA sequences: a case study in the tree genus Milica. Evol Biol 16:259. https://doi.org/10.1186/st12862-016-0831-9
Daïnou K, Flot J-F, Degen B, Blanc-Jolivet C (2017) DNA taxonomy in the timber genus Milica: evidence of unidirectional introgression in the West African contact zone. Tree Genet Genomes.https://doi.org/10.1007/s11295-017-1174-4
Dauby G, Zaiss R, Blach-Overgaard A, Catarino L, Damen T, Deblauwe V et al (2016) Rainbio: a megadatabase of tropical Africa vascular plants distribution. Phyto Keys 74:1–18
de Lafontaine G, Napier JD, Petit RJ, Hu FS (2018) Invoking adaptation to decipher the genetic legacy of past climate change. Ecology 99:1530–1546
Demenou B, Pineiro R, Hardy OJ (2016) Origin and history of the Dahomey Gapseparating West and Central African rainforests: insights from the phylogeography of the legume tree Distemonanthus benthamianus. J Biogeogr 43:1020–1031
Demenou B, Doucet J-L, Hardy O (2017) History of the fragmentation of the African rainforest in the Dahomey Gap: insight from the demographic history of Terminalia superba. Heredity 120:547–561
Duminil J, Heuertz M, Doucet J-L, Bourland N, Cruaud C, Gavory F et al (2010) Cp-DNA based species identification and phylogeography: application to African tropical tree species. Mol Ecol 19:5469–5483. https://doi.org/10.1111/j.1365-294X.2010.04917.x
Duminil J, Brown RP, Ewedje EBK, Mardulyn P, Doucet J-L, Hardy OJ (2013) Large-scale pattern ofgenetic differentiation within African rainforest trees: insights on the roles of ecological gradients and past climate changes on the evolution of Erythrophleum spp. (Fabaceae). BMC Evol Biol 13:1–13
Dupont LM, Jahns S, Marret F, Ning S (2000) Vegetation change in equatorial West Africa: time-slices for the last 150 ka. Palaeogeogr Palaeoclimatol Palaeoecol 155:95–122. https://doi.org/10.1016/S0031-0182(99)00095-4
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Fayolle A, Swaine MD, Bastin J-F, Bourland N, Comiskey J, Dauby G, Doucet J-L, Gillet J-F, Gourlet-Fleury S, Hardy O, Kirunda B, Kouamé FN, Plumptre AJ (2014) Patterns of tree species composition across tropical African forests. J Biogeogr.https://doi.org/10.1111/jbi.12382
Fontaine C, Lovett PN, Sanou H, Maley J, Bouvet J-M (2004) Genetic diversity of the shea tree (Vitellaria paradoxa C.F. Gaertn), detected by RAPD and chloroplast microsatellite markers. Heredity 93:639–648. https://doi.org/10.1038/sj.hdy.6800591
Friedl MA, Sulla-Menashe D, Tan B, Schneider A, Ramankutty N, Sibley A, Huang X (2010) MODIS collection 5 global land cover: algorithm refinements and characterization of new datasets, 2001–2012, collection 5.1 IGBP land cover. Boston University, Boston
Gomez C, Dussert S, Hamon P, Hamon S, Kochko A, Poncet V (2009) Current genetic differentiation of Coffeacanephora Pierre ex A. Froehnin the Guineo-Congolian African zone: cumulative impact of ancient climatic changes and recent human activities. BMC Evol Biol 9:167. https://doi.org/10.1186/1471-2148-9-167
Hardy OJ, Vekemans X (2001) Patterns of allozyme variation in diploid and tetraploid Centaurea jacea at different spatial scales. Evolution 55:943–954
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620. https://doi.org/10.1046/j.1471-8286.2002.00305.x
Hardy OJ, Charbonnel N, Fréville H, Heuertz M (2003) Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163:1467–1482
Hardy OJ, Born C, Budde K, Daïnou K, Dauby G, Duminil J et al (2013) Comparative phylogeography of African rainforest trees: a review of genetic signatures of vegetation history in the Guineo-Congolian region. C R Geosci 345:284–296. https://doi.org/10.1016/j.crte.2013.05.001
Harrison RG, Bogdanowicz (1997) Patterns of variation and linkage disequilibrium in a field cricket hybrid zone. Evolution 51:493. https://doi.org/10.2307/2411122
Hutchinson J, Dalziel JM (1954) Flora of west tropical Africa, 2nd ed. Crown Agents for Overseas Goverments Administrations, London
Ikabanga DU, Stévart T, Koffi GK, Monthe FK, Nzigou Doubindou EC, Dauby G et al (2017) Combining morphology and population genetic analysis uncover species delimitation in the widespread African tree genus Santiria (Burseraceae). Phytotaxa 321:166–180. https://doi.org/10.11646/phytotaxa.321.2.2
IUCN (2014) The IUCN red list of threatened species. Version 2014.3. www.iucnredlist.org. Accessed 25 Sept 2015
Jiggins CD, Mallet J (2000) Biomodal hybrid zones and speciation. Trends Ecol Evol 15:250–255
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jost L, Archer F, Flanagan S, Gaggiotti O, Hoban S, Latch E (2018) Differentiation measures for conservation genetics. Evol Appl 11 (7):1139–1148
Keay RWJ (1953) Revision of the “Flora of west tropical Africa: Ochnaceae. Kew Bull 8:487–492
Koffi KG, Hardy OJ, Doumenge C, Cruaud C, Heuertz M (2011) Diversity gradients and phylogeographic patterns in a widespread African tree typical of mature rainforests, Santiria trimera (Burseraceae). Am J Bot 98:254–264. https://doi.org/10.3732/ajb.1000220
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) CLUMPAK: a programm for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Res 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Letouzey R (1957) La forêt à Lophira alata de la zone littorale camerounaise. Bois For Trop 53:9–20
Letouzey R (1968) Étude phytogéographique du Cameroun. Paris
Li YL, Liu JX (2018) StructureSelector: a web based software to select and visualize the optimal number of clusters using multiple methods. Mol Ecol Res 18:176–177
Lissamou B-J, Couvreur TLP, Atteke C, Stevart T, Piñeiro R, Dauby G et al (2018) Species delimitation in the genus Greenwayodendron based on morphological and genetic markers reveal new species. Taxon 68:442–454
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Lowe AJ, Harris D, Dormontt E, Dawson IK (2010) Testing putative African tropical forest refugia using chloroplast and nuclear DNA phylogeography. Trop Plant Biol 3:50–58. https://doi.org/10.1007/s12042-010-9045-2
Lowry DB, Rockwood RC, Willis JH (2008) Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution 62:2196–2214. https://doi.org/10.1111/j.1558-5646.2008.00457.x
Maley J (1996) The African rainforest: main characteristics of changes in vegetationand climate from the upper cretaceous to quaternary. Proc R Soc Edinb 104B:31–73
Mapongmetsem P-M (2007) Lophira lanceolata Tiegh. ex Keay. [Internet] Record from PROTA4U. van der Vossen HAM. and Mkamilo GS (eds). PROTA (Plant Resources of Tropical Africa/Ressources végétales de l’Afrique tropicale), Wageningen, Netherlands. http://www.prota4u.org/search.as. Accessed 23 Dec 2019
Meunier Q, Moumbogou C, Doucet J-L (2015) Les arbres utiles du Gabon. Presses agronomiques de Gembloux, Gembloux
Migliore J, Kaymak E, Mariac C, Couvreur TLP, Lissambou B-J, Piñeiro R, Hardy OJ (2018) Pre-Pleistocene origin of phylogeographical breaks in African rain forest trees: New insights from (Annonaceae) phylogenomics. J Biogeogr. https://doi.org/10.1111/jbi.13476
Mondol S, Moltke I, Hart J, Keigwin M, Brown L, Stephans et al (2015) New evidence for hybrid zones of forest and savanna elephants in Central and West Africa. Mol Ecol 24:6134–6147. https://doi.org/10.1111/mec.13472
Morales CL, Traveset A (2008) Interspecific pollen transfer: magnitude, prevalence and consequences for plant fitness. Crit Rev Plant Sci 27:221–238. https://doi.org/10.1080/07352680802205631
Morgan-Richards M, Trewick SA, Stringer IA (2010) Geographic parthenogenesis and the common tea-tree stick insect of New Zealand. Mol Ecol 19:1227–1238. https://doi.org/10.1111/j.1365-294X.2010.04542.x
Nei M (1978) Estimation of average heterozygosity and genetic distance froma small number of individuals. Genetics 89:583–590
Odee DW, Telford A, Wilson J, Gaye A, Cavers S (2012) Plio-Pleistocene history and phylogeography of Acacia senegal in dry woodlands and savannahs of sub-Saharan tropical Africa: evidence of early colonisation and recent range expansion. Heredity 109:372–382. https://doi.org/10.1038/hdy.2012.52
Persinos GJ, Quimby MW (1968) Studies on Nigerian plants V. comparative anatomy of Lophira lanceolata and Lophira alata. Econ Bot 22:206–220
Piñeiro R, Staquet A, Hardy OJ (2015) Isolation of nuclear microsatellites in the African timber tree Lophira alata (Ochnaceae) and cross-amplification in L. lanceolata. Appl Plant Sci 3:1500–1056. https://doi.org/10.3732/apps.1500056
Plana V, Gascoigne A, Forrest LL, Harris D, Pennington RT (2004) Pleistocene and pre-pleistocene Begonia speciation in Africa. Mol Phylogenet Evol 31:449–461. https://doi.org/10.1016/j.ympev.2003.08.023
Pritchard JK, Stephens JC, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puechmaille SJ (2016) The program STRUCTURE does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Res 16:608–627. https://doi.org/10.1111/1755-0998.12512
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237. https://doi.org/10.1046/j.1523-1739.2003.01236.x
Rieseberg LH, Carney SE (1998) Plant hybridization. New Phytol 140:599–624
Satabie B (1982) Le phénomène de vicariance chez deux espèces ecophylétiques au Cameroun: Lophira alata Banks ex Gaertn. f. et Lophira lanceolata Van Tiegh. ex Keay (Ochnaceae), PhD thesis, Universite de Yaounde, Cameroon
Schemske DW (2000) Understanding the origin of species. Evolution 54:1069–1073
Sexton GJ, Fere CH, Kalinganire A, Uwamariya A, Lowe AJ, Godwin ID et al (2015) Influence of putative forest refugia and biogeographic barriers on the level and distribution of genetic variation in an African savannah tree, Khaya senegalensis (Desr.) A. Tree Genet Genomes 11:103. https://doi.org/10.1007/s11295-015-0933-3
Smith TB, Schneider CJ, Holder K (2001) Refugial isolation versus ecological gradients. Genetica 112/113:383–398
Sobel JM, Chen GF, Watt LR, Schemske DW (2009) The biology of speciation. Evolution 64:295–315. https://doi.org/10.1111/j.1558-5646.2009.00877.x
Von Thieme HW (1929) Das bongossiholz und seine abstammung. Botanisches Archiv. Band 26. Leipzig
Weir BS (1996) Genetic data analysis. II. Sinauer Associates, Sun-derland, MA
White F (1979) The Guineo-Congolian region and its relationships to other phytochoria. Bull Natl Plant Belg 49:11–55
Whitlock MC (2011) Gst’ and D do not replace Fst. Mol Ecol 20:1083–1091
Acknowledgements
Financial support was provided by the Belgian Federal Science Policy Office (Belspo) through a postdoctoral grant (EEBK) within the project AFRIFORD, and by the F.R.S.-FNRS (Grant No. J.0292.17). Furthermore, this project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Grant Agreement No. 801505. We would like to thank Dakis-Yaoba Ouedraogo for providing herbarium data for L. alata individuals from Gabon including phenological traits, and Marc Sosef for having compared the few herbarium vouchers from western Gabon and eastern/northern Gabon available at the Meise Botanic Garden (BR). We thank CENAREST (Gabon) for the sampling permission (AR0012/18). Moreover, we thank the three anonymous reviewers whose comments have greatly improved this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ewédjè, EE.B.K., Jansen, S., Koffi, G.K. et al. Species delimitation in the African tree genus Lophira (Ochnaceae) reveals cryptic genetic variation. Conserv Genet 21, 501–514 (2020). https://doi.org/10.1007/s10592-020-01265-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-020-01265-7