Abstract
Mitochondria are sensitive to oxidative stress, including that derived from ionizing radiation. To quantify the effects of exposure to environmental radionuclides on mitochondrial DNA (mtDNA) dynamics in wildlife, bank voles (Myodes glareolus) were collected from the chernobyl exclusion zone (CEZ), where animals are exposed to elevated levels of radionuclides, and from uncontaminated areas within the CEZ and elsewhere in Ukraine. Brains of bank voles from outside the CEZ were characterized by low mtDNA copy number and low mtDNA damage; by contrast, bank voles within the CEZ had high mtDNA copy number and high mtDNA damage, consistent with putative damaging effects of elevated radiation and a compensatory response to maintain sufficient functioning mitochondria. In animals outside the CEZ, the expression levels of PGC-1α gene and mtDNA copy number were positively correlated as expected from this gene’s prominent role in mitochondrial biogenesis; this PGC-1α-mtDNA copy number association is absent in samples from the CEZ. Our data imply that exposure to radionuclides is associated with altered mitochondrial dynamics, evident in level of mtDNA and mtDNA damage and the level of activity in mitochondrial synthesis.
Similar content being viewed by others
Introduction
Mitochondria are the powerhouses of eukaryotic cells, involved in energy production, metabolism and cell signaling (Chinnery and Hudson 2013). Cells contain many mitochondria, each of which has multiple copies of its own circular genome (the mitochondrial DNA, mtDNA): mammalian somatic cells may thus contain up to 104 mtDNA copies (Shoubridge and Wai 2007). The number of mitochondria and mtDNA genomes is dynamic and the organelles and their DNA can be replicated or degraded independently of the cell cycle (Michel et al. 2012; Chinnery and Hudson 2013). Reactive oxygen species (ROS) are natural byproducts of mitochondrial function, and indeed, mitochondria present the main source of intracellular ROS. Mitochondrial ROS are typically detoxified by antioxidants or can have important roles, such as cell signaling and immunological defense (Finkel and Holbrook 2000; Shokolenko et al. 2009; Alexeyev et al. 2013). However, damage to mitochondria and/or their genomes can elevate levels of intracellular ROS, leading to greater oxidative stress and cellular damage (Shokolenko et al. 2009; Kam and Banati 2013). Dysfunctional mitochondria and impaired clearance of damaged mitochondria have been associated with various diseases and accelerated molecular aging (Finkel and Holbrook 2000; Correia-Melo et al. 2014).
Mitochondria are thought to be vulnerable to oxidative damage (Finkel and Holbrook 2000; Lee and Wei 2005), as they are suggested to have fewer DNA repair pathways than for nuclear DNA, and the mtDNA is at close proximity to the site of mitochondrial ROS production (Yakes and Van Houten 1997; Stein and Sia 2017). Indeed, oxidative stress induces DNA lesions, such as strand breaks and base modifications, at a higher frequency in mtDNA compared with nuclear DNA (Finkel and Holbrook 2000; Alexeyev et al. 2013). Regulation of the numbers of mitochondria, mtDNA and their integrity is crucial given that excess ROS from damaged mitochondria could positively feedback to cause further mitochondrial damage (and thus more ROS). To mitigate the potentially damaging effects (elevated ROS, inefficient energy production) of dysfunctional mitochondria in response to stress, cells may increase mitochondrial DNA copy number (Kam and Banati 2013), and/or increase transcription of genes associated with mitochondrial biogenesis (Lee and Wei 2005; St-Pierre et al. 2006; Jendrach et al. 2008); notably, peroxisome proliferator-activated receptor gamma coactivator (PGC-1α) is a key regulator of mitochondrial biogenesis, through its role in activating various transcription factors (St-Pierre et al. 2006).
Mitochondria are sensitive to exposure to toxins and other stressors, with the amount of mtDNA damage being a putative biomarker of level of exposure to environment stress; for example, exposure to heavy metals, toxic chemicals or air pollutants (Meyer et al. 2013; Jayasundara 2017; Roubicek and de Souza-Pinto 2017). However, most of our understanding of the effects of toxins on mitochondrial function is derived from laboratory studies (Meyer et al. 2013; Roubicek and de Souza-Pinto 2017) and estimates of levels of mtDNA damage in wildlife are scarce. Nonetheless, mtDNA damage has been detected in bats exposed to mercury contamination (Karouna-Renier et al. 2014), and also in the sun-blistered skin of whales exposed to UV-radiation (Bowman et al. 2013). Additionally, Atlantic killifish exposed to environmental pollutants (PAHs, i.e. polycyclic aromatic hydrocarbons) show higher levels of damage in mtDNA, as well as nuclear DNA in several tissues, with the highest mtDNA damage observed in the brain tissue (Jung et al. 2009). Ionizing radiation is associated with elevated DNA damage (Ward 1988). Under laboratory conditions, exposure to ionizing radiation induces mtDNA damage and mitochondrial dysfunction, leading to elevated levels of mitochondrial ROS production and further oxidative damage to cells (Leach et al. 2001; Murphy et al. 2005; Azzam et al. 2012; Kam and Banati 2013). Thus, exposure to environmental radionuclides is a candidate cause of increased mitochondrial damage. The accident at the former nuclear power plant in Chernobyl, Ukraine (in 1986) released huge amounts of radionuclides over much of Ukraine, Belarus and western Russia, as well as other parts of Europe. Wildlife inhabiting areas contaminated by persistent radionuclides (mostly 137Cs, 90Sr and 239Pu) are exposed to elevated levels of ionizing radiation, and often show detrimental physiological and molecular effects, such as increase in DNA damage, mutation rates and oxidative stress, compromised immune responses and sperm abnormalities (Møller and Mousseau 2015; Einor et al. 2016; Lourenço et al. 2016). Given the high susceptibility to oxidative stress and accumulation of DNA damage, as well as the apparent weaker DNA repair mechanisms available to mitochondria, we hypothesized that exposure to environmental radionuclides impacts mitochondrial homeostasis in wildlife. We examined this hypothesis by measuring (1) mtDNA copy number, (2) amount of damaged mtDNA, and (3) expression of PGC-1α in the brain tissue of bank voles, Myodes glareolus, that had inhabited locations in Ukraine that are contaminated or uncontaminated by radionuclides. We studied brain tissue as it is a metabolically active organ with high mitochondrial copy number, and yet brain tissue has poor mtDNA repair capacity (Karahalil et al. 2002) and is sensitive to oxidative stress during development (Kaindl et al. 2006).
Materials and methods
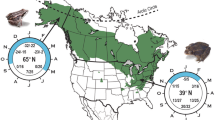
Bank voles were trapped during summer 2015 from contrasting locations within the Chernobyl Exclusion Zone (CEZ), an area of approximately 30 km in radius around the accident site that has restricted access in order to limit human exposure to contamination. Ambient soil background radiation levels were measured at each trap site 1 cm above the ground with a hand-held GM dosimeter (Inspector, International Medcom INC, Sebastopol, CA, USA). Contaminated trapping locations (CEZ elevated, n = 23 voles) within the CEZ were defined by elevated levels of ambient soil radiation (0.46–134 µGy/h) in contrast to the uncontaminated locations (CEZ background, n = 19) (ranging from 0.10–0.28 μGy/h). In bank voles, the external radiation dose of animals at Chernobyl strongly correlates (r = 0.75, Spearman) to the ambient background dose rates measured at the trapping locations (Lavrinienko et al. unpublished). As bank voles may disperse among the mosaic of contaminated and uncontaminated areas within the CEZ, we also sampled animals (control, n = 30) from three uncontaminated locations within Ukraine that were sufficiently far (> 150 km) from the CEZ to ensure that bank voles could not have been exposed to radionuclides (mean background radiation: 0.13 μGy/h) (Fig. 1 and additional details from ESM1).
Map of sampling locations for Myodes glareolus. Location of uncontaminated control sampling sites outside the CEZ (a) and within the chernobyl exclusion zone (CEZ) in Ukraine (b). For the sampling locations within the CEZ (b), ‘CEZ elevated’ locations are marked red circles and the ‘CEZ background’ locations with gray circles. The CEZ is outlined with a dotted line in (b). CNPP chernobyl nuclear power plant
Tissue samples were collected and stored immediately to Allprotect Tissue Reagent (Qiagen). For the mtDNA assays, DNA was extracted from the tissue with DNeasy Blood and Tissue kit (Qiagen). A qPCR assay was developed to estimate the relative mtDNA copy number and the relative amount of undamaged mtDNA copies. The assay is based on amplification of three DNA fragments; a short nuclear gene fragment (N, 36B4 [= Rplp0], the ribosomal phosphoprotein P0, 87 bp), and a short (72 bp) and long (3,252 bp) fragment of mitochondrial genome (see Table S1). Briefly, as polymerase cannot copy damaged DNA, we expect to see a reduction in PCR amplification success of the long mtDNA product because a longer PCR target likely contains one or more sites of strand break/oxidative lesion; by contrast, a small PCR target will rarely incorporate a damaged site and, the short qPCR should reflect the mtDNA copy number (Hunter et al. 2010; Bowman et al. 2013; Zhu and Coffman 2017). The relative copy number estimations were calculated using Pfaffl’s (Pfaffl 2001) formula. In short, the mtDNA copy number in a sample was calculated as a ratio of the short mitochondrial fragment and the nuclear genomic fragment (87 bp fragment of a reference gene 36B4), and ratio of the long mitochondrial fragment and the short mitochondrial fragment was used for the estimation of relative amount undamaged mitochondrial genomes (see ESM1 for full details). Quantitative PCR reactions were performed in 15 µl reactions containing 15 ng of DNA, 200–400 nM of primers (Table S2) and 1 × LightCycler 480 SYBR Green I Master (Roche), using the LightCycler 480 Real-Time PCR System (Roche). Cycling conditions for 36B4 were as follows: 95 °C for 10 min followed by 35 cycles of 95 °C for 10 s, 58 °C for 15 s and 72 °C for 10 s, followed by a melting curve. Cycling conditions for mitochondrial fragments were the same except the quantification step included 35 cycles of 95 °C for 15 s, 57 °C for 15 s and 72 °C for 60 s for the short fragment and 35 cycles of 95 °C for 15 s, 57 °C for 20 s and 72 °C for 3 min for the long fragment. Primers to amplify the short mtDNA fragment were F: 5′-GGGTTGGTAAATTTCGTG-3′, and R: 5′-CGTTTTACGCCGAGAATA-3′ (efficiency 1.92), and for the long mtDNA fragment F: 5′-ATAGTAGAAGCAGGGGCTGGA-3′ and R: 5′-TGGGTTGGAACTAGGCTTGAGT-3′ (efficiency 1.82) (for the nuclear gene 36B4, see primer info in Kesäniemi et al. (2019b). To validate that the assay detects differences in the amount of mtDNA damage, bank vole DNA samples were exposed to direct UV light for increasing amount of time (up to 10 min) to induce mtDNA damage, with control samples without UV exposure. A decreased amplification of the long mtDNA fragment was observed with longer UV exposure times, indicating increased amount of damage (see Fig. S1 and ESM1 for full details).
A subset of samples (n = 39) was used to estimate relative PGC-1α expression among locations. RNA extractions were done using RNeasy Mini Kit (Qiagen) with tissue homogenization using TissueLyser II (Qiagen) and an on-column DNase digestion step. 400 ng of total RNA per sample was used for reverse transcription using iScript cDNA synthesis kit (Bio-Rad). Two reference genes were used in the qPCR reactions, Beta-actin (Actb) and Retention in endoplasmic reticulum sorting receptor 1 (Rer1). Quantitative PCR reactions (qPCRs) for all genes were done using a LightCycler480 Real-Time PCR System (Roche) in a 16 µl final reaction volume that contained ~ 3 ng cDNA template, 0.5 µM of reverse and forward primers and 1X SYBR Green I Master Mix (Roche). Thermal cycling conditions were: 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s, 60 °C for 15 s and 72 °C for 10 s. Primers to amplify PGC-1α were: F: 5′-TGGATGAAGACGGATTGCCC-3′ and R: 5′-AGCTTCTTAAGTAGAGACGGCT-3′ (efficiency 2.07) (see (Jernfors et al. 2018) for primer information for Actb and Rer1, and ESM1 for details of the qPCR protocol and primer design). Relative expression of PGC-1α was analyzed using GenEx v.6.1 (https://genex.gene-quantification.info/), with efficiency correction of the Cq values, and normalization of PGC-1α expression to the harmonic mean of the two reference genes. Relative expression levels were calculated against a standard reference sample run on each plate (Pfaffl 2001). Relative expression data was log10-transformed to normalise the distribution.
We analyzed the effects of sampling location (CEZ elevated, CEZ background, control), sex and size of the animals, i.e. head width, as fixed factors in the model, on (1) mtDNA copy number, (2) amount of damaged mtDNA (where the mtDNA copy number was also added as a factor in the model), and (3) expression of PGC-1α using linear models in R v.3.5.2 (The R Core Team 2018).
Results
Bank voles inhabiting both CEZ locations had significantly more mtDNA copies in their brain tissue compared with animals from the outside the CEZ (Table 1), with the highest mean mtDNA copy number observed in animals from locations of elevated radiation (Fig. 2a, Table S3/ESM1). Moreover, there was significantly greater inter-individual variation in mtDNA copy number in the samples from the CEZ compared with those from the control site (Fig. 2a, Table S3/ESM1, Levene’s F-statistic, F = 15.46, df = 2.71, p < 0.001, see also Fig. S2 for locations outside the CEZ shown separately). High mtDNA copy number was significantly associated with greater damage to mtDNA (Table 1): thus, bank voles from both CEZ locations had significantly fewer undamaged mtDNA copies compared with animals from outside the CEZ (Table 1, Fig. 2b), albeit with a trend of more damage in the CEZ background areas (Table S3/ESM1).
Transcription of PGC-1α was significantly upregulated in the brains of animals collected from the CEZ sites of elevated radiation compared to animals from outside the CEZ. A trend of elevated expression in CEZ background site animals was also seen, however this was not significant (p = 0.066) (Table 1). Consistent with its role in mitochondrial biogenesis, there was a positive correlation between PGC-1α expression and mtDNA copy number (Spearman’s rho, ρ = 0.431, n = 39, p = 0.006). However, when the three sample locations were analyzed separately, this gene expression-copy number association was maintained only in brains of animals from the control locations (ρ = 0.842, n = 14, p < 0.001), whereas weakly negative (CEZ elevated, ρ = − 0.650, n = 12, p = 0.022) or non-significant correlations (CEZ background: ρ = 0.203, n = 13, p = 0.505) were observed in the samples from the CEZ (Fig. 3). No sex effect was seen in any of the analyses (Table 1).
Discussion
Mitochondria provide essential functions to eukaryotic cells, with dynamic changes in mitochondria copy number potentially allowing and contributing to a response to changes in cellular energy demands and/or stress (Michel et al. 2012). Oxidative stress and exposure to ionizing radiation induce mitochondrial dysfunction (Kam and Banati 2013; Meyer et al. 2013), however little is known about the effects of chronic radiation exposure on mitochondrial maintenance in wildlife. We show that exposure to environmental radionuclides impacts mitochondrial homeostasis in the brains of bank voles via an increase in (1) mtDNA copy number and (2) mtDNA damage, and (3) can impact the association between PGC-1α expression and mtDNA copy number.
Exposure to genotoxic stressors such as radiation, pollutants, oxidative stress or DNA damaging chemicals induce an increase in mitochondrial biogenesis (Lin et al. 2013; Correia-Melo et al. 2014; Kaur et al. 2014; de Quadros et al. 2016). In addition, damage to mitochondria can alter mitochondrial function and cellular energy metabolism (Salin et al. 2012; Jayasundara 2017). An increase in mtDNA copy number may be a common response to elevated mtDNA damage derived from exposure to radiation. Human and mouse cells exposed to an acute, high dose of radiation (for example > 3 Gy) increase [by up to threefold (Kam and Banati 2013)] their mtDNA copy number (Malakhova et al. 2005; Kam and Banati 2013; Dannenmann et al. 2017); we show that exposure to environmental radionuclides elicits a comparable response in the brains of bank vole. While increasing the copy number of mitochondria or mtDNA may be needed to maintain sufficient energy production and overall mitochondrial function within cells, this response may further increase the amount of mitochondrial ROS (Lee and Wei 2005), unless there is a concomitant increase in effort to repair or remove the damaged mitochondria/mtDNA. Degradation of damaged mitochondria is therefore a typical response to acute oxidative stress exposure (Shokolenko et al. 2009; Bess et al. 2012) and these pathways also need to be studied to better understand the response to environmental radionuclides, and presumably other stresses, experienced by wildlife. That high mtDNA copy number was coupled with greater mtDNA damage (i.e. lower undamaged mtDNA CN) suggests that inhabiting the CEZ is associated with impaired capability to degrade or repair damaged mtDNA, at least in brain tissue. In general, tissue-specific differences in mitochondrial DNA maintenance influences mtDNA copy number and damage, for example mouse brain tissue with high mitochondrial mass can accumulate high levels of mtDNA damage (Herbers et al. 2019). Additionally, the repair capacity of mtDNA is tissue-specific, with characteristics such as high sensitivity to oxidative stress, slower repair and faster accumulation of oxidative damage observed in mice brains compared with other tissues (testis, kidney and liver) (Karahalil et al. 2002) indicating that the mitochondria in brain tissue could be particularly sensitive to exposure to environment stress. Indeed, brain development is affected by exposure to elevated levels of radiation: prenatal exposure to ionizing radiation is associated with impaired neurogenesis (Tang et al. 2017), as well as reduced brain size in laboratory mice (Verreet et al. 2016), and also in birds (Møller et al. 2011) and bank voles (Kivisaari et al. unpublished) inhabiting contaminated areas in the Chernobyl region.
Increased transcription of PGC-1α in the CEZ samples, where mtDNA copy number was the highest, is consistent with this gene’s key role in regulating mitochondrial biogenesis (St-Pierre et al. 2006; Austin and St-Pierre 2012) and that cells with mtDNA damage should increase mitochondrial biogenesis to maintain sufficient functional mitochondria. However, the loss of the positive correlation between PGC-1α expression and mtDNA copy number in bank voles inhabiting the CEZ may indicate altered regulation of mitochondrial biogenesis or a change in the action of mitochondrial degradation-repair dynamics. Therefore, further experimental studies are needed to more specifically examine the regulation of mitochondrial biogenesis in response to stress within the CEZ. Similarly, a disruption of telomere homeostasis was observed in bank voles inhabiting the CEZ, as the within-individual correlation in telomere length of different tissues was weakened (Kesäniemi et al. 2019b). Moreover, PGC-1α regulates a number of other cellular pathways, including ROS metabolism (St-Pierre et al. 2006; Austin and St-Pierre 2012). Elevated expression of PGC-1α in bank voles inhabiting the CEZ could be directed towards antioxidant pathways to mitigate oxidative stress (Lee and Wei 2005; Valle et al. 2005; St-Pierre et al. 2006; Austin and St-Pierre 2012). Such a response may partly explain the qualitative pattern of slightly lower mtDNA damage and PGC-1α upregulation in animals from the elevated contamination areas compared with those animals from the areas where elevated levels of radionuclides were not detectable within the CEZ.
The numbers of mitochondria and mtDNA genomes within eukaryotic cells and tissues are dynamic (Michel et al. 2012; Meyer et al. 2013; Monternier et al. 2014; Herbers et al. 2019). The generally similar pattern of mtDNA dynamics and PGC-1α expression in samples from the contaminated and uncontaminated areas within the CEZ implied that an animal’s recent exposure to radiation does not elicit a distinct change in mitochondrial dynamics. As the CEZ presents a mosaic of radionuclide contamination over a scale of ~ 1.5 km (Chesser et al. 2004), and bank voles are dispersive, the pattern of elevated levels of mtDNA copy number and mtDNA damage may reflect previous exposure to contaminants in the radioactive landscapes within the CEZ. An increase in the average and the variance in mtDNA copy number and/or damage is expected in a sample of wild animals who have had diverse opportunities (e.g. during foraging events or dispersal to find new territories) to encounter radionuclide contamination during their lifetimes. Current patterns of mitochondrial dynamics may also have been shaped by selection or epigenetic effects in the vole population recolonizing the contaminated CEZ area after the accident (Omar-Nazir et al. 2018). Interestingly, skin fibroblasts isolated from bank voles inhabiting the contaminated area of the CEZ have greater resistance against oxidative and DNA damaging stressors and elevated basal antioxidant levels compared to cells from control voles collected from outside the CEZ (Mustonen et al. 2018). While inhabiting an environment contaminated with radionuclides can induce elevated levels of oxidative stress, DNA damage and mutation rate (Møller and Mousseau 2015; Einor et al. 2016; Lourenço et al. 2016), there is no evidence for an increased rate of accumulation of mutations in the mitochondrial genome, i.e. heteroplasmy, in muscle tissue of bank voles inhabiting the CEZ (Kesäniemi et al. 2018). While changes in gene expression are observed in voles inhabiting the contaminated CEZ areas (Kesäniemi et al. 2019a), the elevated level of genetic variation seen using mitochondrial markers in bank voles within the CEZ is explained by demographic processes rather than exposure to environmental radiation (Meeks et al. 2009). More generally, our data show how human impacts on the environment can affect mitochondrial dynamics in wildlife. Future studies to quantify the heritable or epigenetic components of mtDNA copy number would provide insights into the evolutionary consequences of chronic exposure to environmental contaminants on mitochondrial homeostasis.
Data availability
The datasets (qPCR data) generated during the current study are available from the corresponding author on reasonable request.
References
Alexeyev M, Shokolenko I, Wilson G, LeDoux S (2013) The maintenance of mitochondrial DNA integrity–critical analysis and update. Cold Spring Harb Perspect Biol 5:a012641–a012641. https://doi.org/10.1101/cshperspect.a012641
Austin S, St-Pierre J (2012) PGC1 and mitochondrial metabolism-emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci 125:4963–4971. https://doi.org/10.1242/jcs.113662
Azzam EI, Jay-Gerin JP, Pain D (2012) Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 327:48–60
Bess AS, Crocker TL, Ryde IT, Meyer JN (2012) Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res 40:7916–7931. https://doi.org/10.1093/nar/gks532
Bowman A, Martinez-Levasseur LM, Acevedo-Whitehouse K et al (2013) The simultaneous detection of mitochondrial DNA damage from sun-exposed skin of three whale species and its association with UV-induced microscopic lesions and apoptosis. Mitochondrion 13:342–349. https://doi.org/10.1016/j.mito.2013.04.003
Chesser RK, Bondarkov M, Baker RJ et al (2004) Reconstruction of radioactive plume characteristics along Chernobyl’s Western Trace. J Environ Radioact 71:147–157. https://doi.org/10.1016/S0265-931X(03)00165-6
Chinnery PF, Hudson G (2013) Mitochondrial genetics. Br Med Bull 106:135–159
Correia-Melo C, Hewitt G, Passos JF (2014) Telomeres, oxidative stress and inflammatory factors: partners in cellular senescence? Longev Heal 3:1. https://doi.org/10.1186/2046-2395-3-1
Dannenmann B, Lehle S, Lorscheid S et al (2017) Simultaneous quantification of DNA damage and mitochondrial copy number by long-run DNA-damage quantification (LORD-Q). Oncotarget 8:112417–112425. https://doi.org/10.18632/oncotarget.20112
de Quadros T, Schramm H, Zeni EC et al (2016) Developmental effects of exposure to ultraviolet B radiation on the freshwater prawn Macrobrachium olfersi: Mitochondria as a target of environmental UVB radiation. Ecotoxicol Environ Saf 132:279–287. https://doi.org/10.1016/j.ecoenv.2016.06.018
Einor D, Bonisoli-Alquati A, Costantini D et al (2016) Ionizing radiation, antioxidant response and oxidative damage: a meta-analysis. Sci Total Environ 548–549:463–471. https://doi.org/10.1016/j.scitotenv.2016.01.027
Finkel T, Holbrook NJ (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408:239–247. https://doi.org/10.1038/35041687
Herbers E, Kekäläinen NJ, Hangas A et al (2019) Tissue specific differences in mitochondrial DNA maintenance and expression. Mitochondrion 44:85–92. https://doi.org/10.1016/j.mito.2018.01.004
Hunter SE, Jung D, Di Giulio RT, Meyer JN (2010) The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods 51:444–451. https://doi.org/10.1016/j.ymeth.2010.01.033
Jayasundara N (2017) Ecological significance of mitochondrial toxicants. Toxicology 391:64–74. https://doi.org/10.1016/j.tox.2017.07.015
Jendrach M, Mai S, Pohl S et al (2008) Short- and long-term alterations of mitochondrial morphology, dynamics and mtDNA after transient oxidative stress. Mitochondrion 8:293–304. https://doi.org/10.1016/j.mito.2008.06.001
Jernfors T, Kesäniemi J, Lavrinienko A et al (2018) Transcriptional upregulation of DNA damage response genes in bank voles (Myodes glareolus) inhabiting the chernobyl exclusion zone. Front Environ Sci 5:95. https://doi.org/10.3389/fenvs.2017.00095
Jung D, Cho Y, Collins LB et al (2009) Effects of benzo[a]pyrene on mitochondrial and nuclear DNA damage in Atlantic killifish (Fundulus heteroclitus) from a creosote-contaminated and reference site. Aquat Toxicol 95:44–51. https://doi.org/10.1016/j.aquatox.2009.08.003
Kaindl AM, Sifringer M, Zabel C et al (2006) Acute and long-term proteome changes induced by oxidative stress in the developing brain. Cell Death Differ 13:1097. https://doi.org/10.1038/sj.cdd.4401796
Kam WW-Y, Banati RB (2013) Effects of ionizing radiation on mitochondria. Free Radic Biol Med 65:607–619. https://doi.org/10.1016/j.freeradbiomed.2013.07.024
Karahalil B, Hogue BA, de Souza-Pinto NC, Bohr VA (2002) Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J 16:1895–1902. https://doi.org/10.1096/fj.02-0463com
Karouna-Renier NK, White C, Perkins CR et al (2014) Assessment of mitochondrial DNA damage in little brown bats (Myotis lucifugus) collected near a mercury-contaminated river. Ecotoxicology 23:1419–1429. https://doi.org/10.1007/s10646-014-1284-9
Kaur K, Chauhan V, Gu F, Chauhan A (2014) Bisphenol A induces oxidative stress and mitochondrial dysfunction in lymphoblasts from children with autism and unaffected siblings. Free Radic Biol Med 76:25–33. https://doi.org/10.1016/j.freeradbiomed.2014.07.030
Kesäniemi J, Boratyński Z, Danforth J et al (2018) Analysis of heteroplasmy in bank voles inhabiting the Chernobyl exclusion zone: a commentary on Baker et al. (2017) “Elevated mitochondrial genome variation after 50 generations of radiation exposure in a wild rodent”. Evol Appl 11:820–826
Kesäniemi J, Jernfors T, Lavrinienko A et al (2019a) Exposure to environmental radionuclides is associated with altered metabolic and immunity pathways in a wild rodent. Mol Ecol. 28:4620–4635. https://doi.org/10.1111/mec.15241
Kesäniemi J, Lavrinienko A, Tukalenko E et al (2019b) Exposure to environmental radionuclides associates with tissue-specific impacts on telomerase expression and telomere length. Sci Rep. 9(1):850. https://doi.org/10.1038/s41598-018-37164-8
Leach JK, Van Tuyle G, Lin PS et al (2001) Ionizing radiation-induced, mitochondria-dependent generation of reactive oxygen/nitrogen. Cancer Res 61:3894–3901
Lee H-C, Wei Y-H (2005) Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol 37:822–834. https://doi.org/10.1016/j.biocel.2004.09.010
Lin Y, Sun X, Qiu L et al (2013) Exposure to bisphenol A induces dysfunction of insulin secretion and apoptosis through the damage of mitochondria in rat insulinoma (INS-1) cells. Cell Death Dis 4(1):e460. https://doi.org/10.1038/cddis.2012.206
Lourenço J, Mendo S, Pereira R (2016) Radioactively contaminated areas: Bioindicator species and biomarkers of effect in an early warning scheme for a preliminary risk assessment. J Hazard Mater 317:503–542
Malakhova L, Bezlepkin VG, Antipova V et al (2005) The increase in mitochondrial DNA copy number in the tissues of Γ-irradiated mice. Cell Mol Biol Lett 10:721–732
Meeks HN, Chesser RK, Rodgers BE et al (2009) Understanding the genetic consequences of environmental toxicant exposure: Chernobyl as a model system. Environ Toxicol Chem 28:1982–1994. https://doi.org/10.1897/08-578.1
Meyer JN, Leung MCK, Rooney JP et al (2013) Mitochondria as a target of environmental toxicants. Toxicol Sci 134:1–17
Michel S, Wanet A, De Pauw A et al (2012) Crosstalk between mitochondrial (dys)function and mitochondrial abundance. J Cell Physiol 227:2297–2310
Møller AP, Mousseau TA (2015) Strong effects of ionizing radiation from Chernobyl on mutation rates. Sci Rep 5:8363. https://doi.org/10.1038/srep08363
Møller AP, Bonisoli-Alquati A, Rudolfsen G, Mousseau TA (2011) Chernobyl birds have smaller brains. PLoS ONE 6:e16862. https://doi.org/10.1371/journal.pone.0016862
Monternier P-A, Marmillot V, Rouanet J-L, Roussel D (2014) Mitochondrial phenotypic flexibility enhances energy savings during winter fast in king penguin chicks. J Exp Biol 217:2691–2697. https://doi.org/10.1242/jeb.104505
Murphy JEJ, Nugent S, Seymour C, Mothersill C (2005) Mitochondrial DNA point mutations and a novel deletion induced by direct low-LET radiation and by medium from irradiated cells. Mutat Res Genet Toxicol Environ Mutagen 585:127–136. https://doi.org/10.1016/j.mrgentox.2005.04.011
Mustonen V, Kesäniemi J, Lavrinienko A et al (2018) Fibroblasts from bank voles inhabiting Chernobyl have increased resistance against oxidative and DNA stresses. BMC Cell Biol 19:17
Omar-Nazir L, Shi X, Møller A et al (2018) Long-term effects of ionizing radiation after the Chernobyl accident: possible contribution of historic dose. Environ Res 165:55–62. https://doi.org/10.1016/j.envres.2018.04.005
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:45e–45. https://doi.org/10.1093/nar/29.9.e45
Roubicek DA, de Souza-Pinto NC (2017) Mitochondria and mitochondrial DNA as relevant targets for environmental contaminants. Toxicology 391:100–108. https://doi.org/10.1016/j.tox.2017.06.012
Salin K, Luquet E, Rey B et al (2012) Alteration of mitochondrial efficiency affects oxidative balance, development and growth in frog (Rana temporaria) tadpoles. J Exp Biol 215:863–869. https://doi.org/10.1242/jeb.062745
Shokolenko I, Venediktova N, Bochkareva A et al (2009) Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res 37:2539–2548. https://doi.org/10.1093/nar/gkp100
Shoubridge E, Wai T (2007) Mitochondrial DNA and the mammalian oocyte. Curr Top Dev Biol 77:87–111. https://doi.org/10.1016/S0070-2153(06)77004-1
Stein A, Sia EA (2017) Mitochondrial DNA repair and damage tolerance. Front Biosci (Landmark Ed) 22:920–943
St-Pierre J, Drori S, Uldry M et al (2006) Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127:397–408. https://doi.org/10.1016/j.cell.2006.09.024
Tang FR, Loke WK, Khoo BC (2017) Low-dose or low-dose-rate ionizing radiation-induced bioeffects in animal models. J Radiat Res 58:165–182. https://doi.org/10.1093/jrr/rrw120
The R Core Team (2018) R: a language and environment for statistical computing.
Valle I, Álvarez-Barrientos A, Arza E et al (2005) PGC-1α regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc Res 66:562–573. https://doi.org/10.1016/j.cardiores.2005.01.026
Verreet T, Verslegers M, Quintens R et al (2016) Current evidence for developmental, structural, and functional brain defects following prenatal radiation exposure. Neural Plast. https://doi.org/10.1155/2016/1243527
Ward JF (1988) DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol 35:95–125. https://doi.org/10.1016/S0079-6603(08)60611-X
Yakes FM, Van Houten B (1997) Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA 94:514–519
Zhu S, Coffman JA (2017) Simple and fast quantification of DNA damage by real-time PCR, and its application to nuclear and mitochondrial DNA from multiple tissues of aging zebrafish. BMC Res Notes 10(1):269. https://doi.org/10.1186/s13104-017-2593-x
Acknowledgements
Open access funding provided by University of Oulu including Oulu University Hospital. We acknowledge Gennadi Milinevsky, Igor Chizhevsky, Andrii Simon, Serhii Kirieiev, Anatoly Nosovsky and Maksym Ivanenko for logistic support and help in organizing fieldwork in Ukraine. PCW and TM were funded by Academy of Finland (Project Numbers 287153 and 268670). TAM and APM were supported by awards from the Samuel Freeman Charitable Trust and the American Council of Learned Societies. The authors declare no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Human and animal rights
All procedures complied with the legal requirements and international guidelines for the use of animals in research. Permissions for the field experiments and animal handling were obtained from the Finnish Animal Experimentation Committee (Permission Number ESAVI/7256/04.10.07/2014).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10682_2019_10028_MOESM1_ESM.pdf
ESM1. Additional analysis details and the qPCR protocols for mitochondrial assay and gene expression analysis. (PDF 659 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kesäniemi, J., Lavrinienko, A., Tukalenko, E. et al. Exposure to environmental radionuclides alters mitochondrial DNA maintenance in a wild rodent. Evol Ecol 34, 163–174 (2020). https://doi.org/10.1007/s10682-019-10028-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10682-019-10028-x