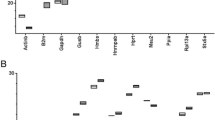
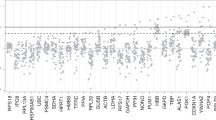
Abstract
Murine bone marrow-derived macrophages (M0) and M1- and M2-polarized macrophages are being widely used as a laboratory model for polarized macrophages related molecular mechanism analysis. Gene expression analysis based on reference gene normalization using RT-qPCR was a powerful way to explore the molecular mechanism. But little is known about reference genes in these cell models. So, the goal of this study was to identify reference genes in these types of macrophages. Candidate reference genes in murine bone marrow-derived and polarized macrophages were selected from microarray data using Limma linear model method and evaluated by determining the stability value using five algorithms: BestKeeper, NormFinder, GeNorm, Delta CT method, and RefFinder. Finally, the selected stable reference genes were validated by testing three important immune and inflammatory genes (NLRP1, IL-1β, and TNF-α) in the cell lines. Our study has clearly shown that Ubc followed by Eef1a1 and B2m respectively were recognized as the three ideal reference genes for gene expression analysis in murine bone marrow-derived and polarized macrophages. When three reference genes with strong different stability were used for validation, a large variation of a gene expression level of IL-1β, TNF-α and NLRP1 were obtained which provides clear evidence of the need for careful selection of reference genes for RT-qPCR analysis. Normalization of mRNA expression level with Ubc rather than Actb or Gusb by qPCR in macrophages and polarized macrophages is required to ensure the accuracy of the qPCR analysis.




Similar content being viewed by others
Data availability
The data and materials used to support the findings in this study are included in the article.
Abbreviations
- BMDM or M0:
-
Bone marrow-derived macrophage
- RT-qPCR:
-
Real-time quantitative PCR
- M1:
-
Classically activated macrophages
- M2:
-
Alternatively activated macrophages
- RG:
-
Reference gene
- STDEV:
-
Standard deviation
References
Davies LC, Taylor PR (2015) Tissue-resident macrophages: then and now. Immunology 144(4):541–548. https://doi.org/10.1111/imm.12451
Wynn TA, Chawla A, Pollard JW (2013) Macrophage biology in development, homeostasis and disease. Nature 496(7446):445–455. https://doi.org/10.1038/nature12034
Navegantes KC, de Souza GR, Pereira PAT, Czaikoski PG, Azevedo CHM, Monteiro MC (2017) Immune modulation of some autoimmune diseases: the critical role of macrophages and neutrophils in the innate and adaptive immunity. J Transl Med 15(1):36. https://doi.org/10.1186/s12967-017-1141-8
Vannella KM, Wynn TA (2017) Mechanisms of organ injury and repair by macrophages. Annu Rev Physiol 79:593–617. https://doi.org/10.1146/annurev-physiol-022516-034356
Isidro RA, Appleyard CB (2016) Colonic macrophage polarization in homeostasis, inflammation, and cancer. Am J Physiol Gastrointest Liver Physiol 311(1):G59–73. https://doi.org/10.1152/ajpgi.00123.2016
Gordon S, Martinez-Pomares L (2017) Physiological roles of macrophages. Pflugers Arch 469(3–4):365–374. https://doi.org/10.1007/s00424-017-1945-7
Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A (2018) Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol 233(9):6425–6440. https://doi.org/10.1002/jcp.26429
Zhou D, Huang C, Lin Z, Zhan S, Kong L, Fang C, Li J (2014) Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal 26(2):192–197. https://doi.org/10.1016/j.cellsig.2013.11.004
Ying W, Cheruku PS, Bazer FW, Safe SH, Zhou B (2013) Investigation of macrophage polarization using bone marrow derived macrophages. J Vis Exp. https://doi.org/10.3791/50323
Koh YC, Yang G, Lai CS, Weerawatanakorn M, Pan MH (2018) Chemopreventive effects of phytochemicals and medicines on M1/M2 polarized macrophage role in inflammation-related diseases. Int J Mol Sci. https://doi.org/10.3390/ijms19082208
Sica A, Mantovani A (2012) Macrophage plasticity and polarization: in vivo veritas. J Clin Investig 122(3):787–795. https://doi.org/10.1172/JCI59643
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25(2):169–193
Appukuttan B, Ashander LM, Ma Y, Smith JR (2018) Selection of reference genes for studies of human retinal endothelial cell gene expression by reverse transcription-quantitative real-time polymerase chain reaction. Gene Rep 10:123–134. https://doi.org/10.1016/j.genrep.2017.11.009
Abuna RPF, Oliveira FS, Ramos JIR, Lopes HB, Freitas GP, Souza ATP, Beloti MM, Rosa AL (2018) Selection of reference genes for quantitative real-time polymerase chain reaction studies in rat osteoblasts. J Cell Physiol. https://doi.org/10.1002/jcp.26886
Tanaka A, To J, O'Brien B, Donnelly S, Lund M (2017) Selection of reliable reference genes for the normalisation of gene expression levels following time course LPS stimulation of murine bone marrow derived macrophages. BMC Immunol 18(1):43. https://doi.org/10.1186/s12865-017-0223-y
Ferraz FB, Fernandez JH (2016) Selection and validation of reference house-keeping genes in the J774A1 macrophage cell line for quantitative real-time PCR. Genet Mol Res 15(1):15017720. https://doi.org/10.4238/gmr.15017720
Stephens AS, Stephens SR, Morrison NA (2011) Internal control genes for quantitative RT-PCR expression analysis in mouse osteoblasts, osteoclasts and macrophages. BMC Res Notes 4:410. https://doi.org/10.1186/1756-0500-4-410
Maess MB, Sendelbach S, Lorkowski S (2010) Selection of reliable reference genes during THP-1 monocyte differentiation into macrophages. BMC Mol Biol 11:90. https://doi.org/10.1186/1471-2199-11-90
Taylor DL, Thomson PC, de Silva K, Whittington RJ (2007) Validation of endogenous reference genes for expression profiling of RAW264.7 cells infected with Mycobacterium avium subsp. paratuberculosis by quantitative PCR. Vet Immunol Immunopathol 115(1–2):43–55. https://doi.org/10.1016/j.vetimm.2006.10.007
Ju W, Smith AO, Sun T, Zhao P, Jiang Y, Liu L, Zhang T, Qi K, Qiao J, Xu K, Zeng L (2018) Validation of housekeeping genes as reference for reverse-transcription-qPCR analysis in busulfan-injured microvascular endothelial cells. Biomed Res Int 2018:4953806. https://doi.org/10.1155/2018/4953806
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Can Res 64(15):5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):0034
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–excel-based tool using pair-wise correlations. Biotechnol Lett 26(6):509–515
Rao X, Huang X, Zhou Z, Lin X (2013) An improvement of the 2(-∆∆CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinform Biomath 3(3):71–85
Panina Y, Germond A, Masui S, Watanabe TM (2018) Validation of common housekeeping genes as reference for qPCR gene expression analysis during iPS reprogramming process. Sci Rep 8(1):8716. https://doi.org/10.1038/s41598-018-26707-8
Zyzynska-Granica B, Koziak K (2012) Identification of suitable reference genes for real-time PCR analysis of statin-treated human umbilical vein endothelial cells. PLoS ONE 7(12):e51547. https://doi.org/10.1371/journal.pone.0051547
Weischenfeldt J, Porse B (2008) Bone marrow-derived macrophages (BMM): isolation and applications. CSH Protoc. https://doi.org/10.1101/pdb.prot5080
Luo Y, Shao L, Chang J, Feng W, Liu YL, Cottler-Fox MH, Emanuel PD, Hauer-Jensen M, Bernstein ID, Liu L, Chen X, Zhou J, Murray PJ, Zhou D (2018) M1 and M2 macrophages differentially regulate hematopoietic stem cell self-renewal and ex vivo expansion. Blood Adv 2(8):859–870. https://doi.org/10.1182/bloodadvances.2018015685
Zhang X, Goncalves R, Mosser DM (2008) The isolation and characterization of murine macrophages. Curr Protoc Immunol. https://doi.org/10.1002/0471142735.im1401s83
Zhou Y, Yoshida S, Kubo Y, Yoshimura T, Kobayashi Y, Nakama T, Yamaguchi M, Ishikawa K, Oshima Y, Ishibashi T (2017) Different distributions of M1 and M2 macrophages in a mouse model of laser-induced choroidal neovascularization. Mol Med Rep 15(6):3949–3956. https://doi.org/10.3892/mmr.2017.6491
Glass EB, Masjedi S, Dudzinski SO, Wilson AJ, Duvall CL, Yull FE, Giorgio TD (2019) Optimizing mannose "click" conjugation to polymeric nanoparticles for targeted siRNA delivery to human and murine macrophages. ACS Omega 4(16):16756–16767. https://doi.org/10.1021/acsomega.9b01465
Laban H, Weigert A, Zink J, Elgheznawy A, Schurmann C, Gunther L, Abdel Malik R, Bothur S, Wingert S, Bremer R, Rieger MA, Brune B, Brandes RP, Fleming I, Benz PM (2018) VASP regulates leukocyte infiltration, polarization, and vascular repair after ischemia. J Cell Biol 217(4):1503–1519. https://doi.org/10.1083/jcb.201702048
Jablonski KA, Amici SA, Webb LM, Ruiz-Rosado Jde D, Popovich PG, Partida-Sanchez S, Guerau-de-Arellano M (2015) Novel markers to delineate murine M1 and M2 macrophages. PLoS ONE 10(12):e0145342. https://doi.org/10.1371/journal.pone.0145342
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Liu Y, Niu M, Yao C, Hai Y, Yuan Q, Liu Y, Guo Y, Li Z, He Z (2015) Fractionation of human spermatogenic cells using STA-PUT gravity sedimentation and their miRNA profiling. Sci Rep 5:8084. https://doi.org/10.1038/srep08084
Velada I, Ragonezi C, Arnholdt-Schmitt B, Cardoso H (2014) Reference genes selection and normalization of oxidative stress responsive genes upon different temperature stress conditions in Hypericum perforatum L. PLoS ONE 9(12):e115206. https://doi.org/10.1371/journal.pone.0115206
Khan S, Roberts J, Wu SB (2017) Reference gene selection for gene expression study in shell gland and spleen of laying hens challenged with infectious bronchitis virus. Sci Rep 7(1):14271. https://doi.org/10.1038/s41598-017-14693-2
Feng X, Xiong Y, Qian H, Lei M, Xu D, Ren Z (2010) Selection of reference genes for gene expression studies in porcine skeletal muscle using SYBR green qPCR. J Biotechnol 150(3):288–293. https://doi.org/10.1016/j.jbiotec.2010.09.949
Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7:33. https://doi.org/10.1186/1471-2199-7-33
Zhao DJ, Guo K, Kang L (2012) Identification of condition-specific reference genes from microarray data for locusts exposed to hypobaric hypoxia. FEBS Open Bio 2:235–240. https://doi.org/10.1016/j.fob.2012.08.001
Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH, Huang CY (2007) Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics 8:140. https://doi.org/10.1186/1471-2164-8-140
Gu YR, Li MZ, Zhang K, Chen L, Jiang AA, Wang JY, Li XW (2011) Evaluation of endogenous control genes for gene expression studies across multiple tissues and in the specific sets of fat- and muscle-type samples of the pig. J Anim Breed Genet 128(4):319–325. https://doi.org/10.1111/j.1439-0388.2011.00920.x
Hoang VLT, Tom LN, Quek XC, Tan JM, Payne EJ, Lin LL, Sinnya S, Raphael AP, Lambie D, Frazer IH, Dinger ME, Soyer HP, Prow TW (2017) RNA-seq reveals more consistent reference genes for gene expression studies in human non-melanoma skin cancers. PeerJ 5:e3631. https://doi.org/10.7717/peerj.3631
Carmona R, Arroyo M, Jimenez-Quesada MJ, Seoane P, Zafra A, Larrosa R, Alche JD, Claros MG (2017) Automated identification of reference genes based on RNA-seq data. Biomed Eng 16(Suppl 1):65. https://doi.org/10.1186/s12938-017-0356-5
Zhan C, Zhang Y, Ma J, Wang L, Jiang W, Shi Y, Wang Q (2014) Identification of reference genes for qRT-PCR in human lung squamous-cell carcinoma by RNA-Seq. Acta Biochim Biophys Sin 46(4):330–337. https://doi.org/10.1093/abbs/gmt153
Funding
This study was supported in part by National Natural Science Foundation of China under Grant [Nos. 81700178, 31872795 and 81570096]; Natural Science Foundation of Jiangsu Province under Grant [No. BK20170259]; Jiangsu Provincial Key Research and Development Program under Grant [No. BE2018637]; Jiangsu Province’s Key Provincial Talents Program under Grant [No. ZDRCA2016054]; China Postdoctoral Science Foundation Grant [No. 2018M632380]; and Jiangsu Postdoctoral Science Foundation under Grant [No. 1701064B] for purchasing reagents, testing fee, and publication charge.
Author information
Authors and Affiliations
Contributions
Conceptualization, AOS; Data curation, WJ and AOS; Formal analysis, KQ; Funding acquisition, WJ; Investigation, WJ, TS, WL, KQ and LZ; Methodology, TS, WL, YB, SYA and KQ; Project administration, WL; Resources, JQ and LZ; Software, TS, WL, YB and SYA; Supervision, LZ; Validation, WJ, KX, JQ and LZ; Visualization, JQ and LZ; Writing—original draft, WJ; Writing—review & editing, AOS, SYA, KX, JQ and LZ.
Corresponding authors
Ethics declarations
Conflict of interest
All authors have no dispute of interest to disclose.
Consent to publish
All authors approved the submission of the manuscript.
Ethics approval and consent to participate
All animal care and procedures were per the ethical standards, approved by Jiangsu Society for Animal Welfare, China (Acceptance number: XZMC20130226) and Science and Technology Department of Jiangsu Province, China (Acceptance number: SYXK(SU)-2015-0030 and SCXK(SU)-2015-0009).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ju, W., Sun, T., Lu, W. et al. Reference gene selection and validation for mRNA expression analysis by RT-qPCR in murine M1- and M2-polarized macrophage. Mol Biol Rep 47, 2735–2748 (2020). https://doi.org/10.1007/s11033-020-05372-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05372-z