Abstract
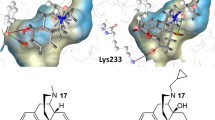
Targeting the mu opioid receptor (MOR) by applying orthosteric ligands is the most frequently employed method to treat opioid use disorder (OUD). Unfortunately, most of MOR orthosteric ligands produce severe side effects, mainly due to their low selectivity over other opioid receptors. In contrast, some G protein-coupled receptor allosteric modulators have been reported to exhibit high subtype selectivity and can effectively modulate the potency and/or efficacy of orthosteric ligands. Recently, NAQ and its analog NCQ were identified as novel MOR bitopic modulators. Interestingly, NAQ and NCQ were similar in structure but exhibited different efficacy profiles to the MOR. NAQ exhibited an antagonism activity to the MOR while NCQ showed a partial agonism activity to the MOR. In the present study, molecular modeling methods were applied to explore the putative molecular mechanisms of their different functional profiles to the MOR. When NAQ binding with the inactive MOR, the ‘address’ portion of NAQ interacted with the MOR allosteric site but showed no significant allosteric modulation of the efficacy of the ‘message’ portion of NAQ. However, when NCQ binding with the inactive and active MOR, the ‘address’ portion of NCQ seemed to be able to positively modulate the efficacy of the ‘message’ portion of NCQ at varying levels. Evidentially, the substituents at the 1′- and 4′-positions of the isoquinoline ring of NCQ seemed to play a critical role in the modulatory function of the ‘address’ portion of NCQ. These findings will be invaluable to develop our next generation of MOR bitopic modulators with high affinity and subtype selectivity to potentially treat OUD.






Similar content being viewed by others
References
National Institute on Drug Abuse (2019) Opioid overdose crisis. https://www.drugabuse.gov/drugs-abuse/opioids/opioid-overdose-crisis. Accessed Jan 2019
Alderks CE (2017) Trends in the use of methadone, buprenorphine, and extended-release naltrexone at substance abuse treatment facilities: 2003–2015 (update) The CBHSQ report: August 22, 2017. Center for Behavioral Health Statistics and Quality Substance Abuse and Mental Health Services Administration, Rockville, MD
Gray A (2007) Department of Therapeutics and Medicines Center for the AIDS Programme of Research in South Africa Congella, South Africa
Matthes HW, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, Befort K, Dierich A, Le Meur M, Dollé P (1996) Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the µ-opioid-receptor gene. Nature 383:819
Law P-Y, Wong YH, Loh HH (2000) Molecular mechanisms and regulation of opioid receptor signaling. Annu Rev Pharmacol Toxicol 40:389–430
Biggio G, Casu M, Corda MG, Di Bello C, Gessa G (1978) Stimulation of dopamine synthesis in caudate nucleus by intrastriatal enkephalins and antagonism by naloxone. Science 200:552–554
Wood PL, Stotland M, Richard J, Rackham A (1980) Actions of mu, kappa, sigma, delta and agonist/antagonist opiates on striatal dopaminergic function. J Pharmacol Exp Ther 215:697–703
Spanagel R, Herz A, Shippenberg TS (1992) Opposing tonically active endogenous opioid systems modulate the mesolimbic dopaminergic pathway. Proc Natl Acad Sci USA 89:2046–2050
Haddou TB, Béni S, Hosztafi S, Malfacini D, Calo G, Schmidhammer H, Spetea M (2014) Pharmacological investigations of N-substituent variation in morphine and oxymorphone: opioid receptor binding, signaling and antinociceptive activity. PLoS ONE 9:e99231
Huang P, Kehner GB, Cowan A, Liu-Chen L-Y (2001) Comparison of pharmacological activities of buprenorphine and norbuprenorphine: norbuprenorphine is a potent opioid agonist. J Pharmacol Exp Ther 297:688–695
Katritch V, Cherezov V, Stevens RC (2013) Structure-function of the G protein–coupled receptor superfamily. Annu Rev Pharmacool Toxicol 53:531–556
Feng Z, Hu G, Ma S, Xie X-Q (2015) Computational advances for the development of allosteric modulators and bitopic ligands in G protein-coupled receptors. AAPS J 17:1080–1095
Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V (2010) Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 330:1091–1095
Burger WA, Sexton PM, Christopoulos A, Thal DM (2018) Toward an understanding of the structural basis of allostery in muscarinic acetylcholine receptors. J Gen Physiol 150:1360–1372
Melancon BJ, Hopkins CR, Wood MR, Emmitte KA, Niswender CM, Christopoulos A, Conn PJ, Lindsley CW (2012) Allosteric modulation of seven transmembrane spanning receptors: theory, practice, and opportunities for central nervous system drug discovery. J Med Chem 55:1445–1464
Yanamala N, Klein-Seetharaman J (2010) Allosteric modulation of G protein coupled receptors by cytoplasmic, transmembrane and extracellular ligands. Pharmaceuticals 3:3324–3332
Marmolejo-Valencia A, Martínez-Mayorga K (2017) Allosteric modulation model of the mu opioid receptor by herkinorin, a potent not alkaloidal agonist. J Comput Aided Mol Des 31:467–482
Cooke R, Congreve M (2016) Allosteric binding: structures reveal new ways to tame G protein-coupled receptors. Future Sci. https://doi.org/10.4155/fmc-2016-0167
Wess J (2005) Allosteric binding sites on muscarinic acetylcholine receptors. Mol Pharmacol 68:1506–1509
Eglen R, Reisine T (2011) GPCRs revisited: new insights lead to novel drugs. Pharmaceuticals 4:244–272
Christopoulos A, Changeux J-P, Catterall WA, Fabbro D, Burris TP, Cidlowski JA, Olsen RW, Peters JA, Neubig RR, Pin J-P (2014) International Union of Basic and Clinical Pharmacology. XC. Multisite pharmacology: recommendations for the nomenclature of receptor allosterism and allosteric ligands. Pharmacol Rev 66:918–947
Tränkle C, Dittmann A, Schulz U, Weyand O, Buller S, Jöhren K, Heller E, Birdsall NJ, Holzgrabe U, Ellis J (2005) Atypical muscarinic allosteric modulation: cooperativity between modulators and their atypical binding topology in muscarinic M2 and M2/M5 chimeric receptors. Mol Pharmacol 68:1597–1610
Ivetac A, Andrew McCammon J (2010) Mapping the druggable allosteric space of G-protein coupled receptors: a fragment-based molecular dynamics approach. Chem Biol Drug Des 76:201–217
Sun B, Bachhawat P, Chu ML-H, Wood M, Ceska T, Sands ZA, Mercier J, Lebon F, Kobilka TS, Kobilka BK (2017) Crystal structure of the adenosine A2A receptor bound to an antagonist reveals a potential allosteric pocket. Proc Natl Acad Sci USA 114:2066–2071
Korczynska M, Clark MJ, Valant C, Xu J, Von Moo E, Albold S, Weiss DR, Torosyan H, Huang W, Kruse AC (2018) Structure-based discovery of selective positive allosteric modulators of antagonists for the M2 muscarinic acetylcholine receptor. Proc Natl Acad Sci USA 115:E2419–E2428
Das D, Maeda K, Hayashi Y, Gavande N, Desai DV, Chang SB, Ghosh AK, Mitsuya H (2015) Insights into the mechanism of inhibition of CXCR4: identification of Piperidinylethanamine analogs as anti-HIV-1 inhibitors. Antimicrob Agents Chemother 59:1895–1904
Garcia-Perez J, Rueda P, Alcami J, Rognan D, Arenzana-Seisdedos F, Lagane B, Kellenberger E (2011) Allosteric model of maraviroc binding to CC chemokine receptor 5 (CCR5). J Biol Chem 286:33409–33421
Ferruz N, Doerr S, Vanase-Frawley MA, Zou Y, Chen X, Marr ES, Nelson RT, Kormos BL, Wager TT, Hou X (2018) Dopamine D3 receptor antagonist reveals a cryptic pocket in aminergic GPCRs. Sci Rep 8:897
Che T, Majumdar S, Zaidi SA, Ondachi P, McCorvy JD, Wang S, Mosier PD, Uprety R, Vardy E, Krumm BE (2018) Structure of the nanobody-stabilized active state of the kappa opioid receptor. Cell 172:55–67
Livingston KE, Stanczyk MA, Burford NT, Alt A, Canals M, Traynor JR (2018) Pharmacologic evidence for a putative conserved allosteric site on opioid receptors. Mol Pharmacol 93:157–167
Huang W, Manglik A, Venkatakrishnan A, Laeremans T, Feinberg EN, Sanborn AL, Kato HE, Livingston KE, Thorsen TS, Kling RC (2015) Structural insights into µ-opioid receptor activation. Nature 524:315–321
Ballesteros JA, Weinstein H (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci 25:366–428
Livingston KE, Traynor JR (2018) Allostery at opioid receptors: modulation with small molecule ligands. Br J Pharmacol 175:2846–2856
Livingston KE (2016) Allosteric Modulation of the Mu Opioid Receptor (Doctoral dissertation).
Burford NT, Watson J, Bertekap R, Alt A (2011) Strategies for the identification of allosteric modulators of G-protein-coupled receptors. Biochem Pharmacol 81:691–702
Conn PJ, Christopoulos A, Lindsley CW (2009) Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov 8:41–54
Bartuzi D, Kaczor AA, Matosiuk D (2016) Interplay between two allosteric sites and their influence on agonist binding in human μ opioid receptor. J Chem Inf Model 56:563–570
Burford NT, Clark MJ, Wehrman TS, Gerritz SW, Banks M, O’Connell J, Traynor JR, Alt A (2013) Discovery of positive allosteric modulators and silent allosteric modulators of the μ-opioid receptor. Proc Natl Acad Sci USA 110:10830–10835
Kathmann M, Flau K, Redmer A, Tränkle C, Schlicker E (2006) Cannabidiol is an allosteric modulator at mu-and delta-opioid receptors. Naunyn Schmiedebergs Arch Pharmacol 372:354–361
Rothman RB, Murphy DL, Xu H, Godin JA, Dersch CM, Partilla JS, Tidgewell K, Schmidt M, Prisinzano TE (2007) Salvinorin A: allosteric interactions at the μ-opioid receptor. J Pharmacol Exp Ther 320:801–810
Reinecke BA, Wang H, Zhang Y (2019) Recent Advances in the Drug Discovery and Development of Dualsteric/Bitopic Activators of G Protein-Coupled Receptors. Curr Top Med Chem 19:2378–2392
Chen X, Klöckner J, Holze J, Zimmermann C, Seemann WK, Schrage R, Bock A, Mohr K, Tränkle C, Holzgrabe U (2014) Rational design of partial agonists for the muscarinic m1 acetylcholine receptor. J Med Chem 58:560–576
Newman AH, Grundt P, Cyriac G, Deschamps JR, Taylor M, Kumar R, Ho D, Luedtke RR (2009) N-(4-(4-(2, 3-dichloro-or 2-methoxyphenyl) piperazin-1-yl) butyl) heterobiarylcarboxamides with functionalized linking chains as high affinity and enantioselective D3 receptor antagonists. J Med Chem 52:2559–2570
Sanna MG, Vincent KP, Repetto E, Nguyen N, Brown SJ, Abgaryan L, Riley SW, Leaf NB, Cahalan SM, Kiosses WB (2016) Bitopic sphingosine 1-phosphate receptor 3 (S1P3) antagonist rescue from complete heart block: pharmacological and genetic evidence for direct S1P3 regulation of mouse cardiac conduction. Mol Pharmacol 89:176–186
Falls BA, Zhang Y (2019) Insights into the allosteric mechanism of setmelanotide (RM-493) as a potent and first-in-class melanocortin-4 receptor (MC4R) agonist to treat rare genetic disorders of obesity through an in silico approach. ACS Chem Neurosci 10:1055–1065
Kamal M, Jockers R (2009) Bitopic ligands: all-in-one orthosteric and allosteric. F1000 Biol Rep. https://doi.org/10.3410/B1-77
Fronik P, Gaiser BI, Sejer Pedersen D (2017) Bitopic ligands and metastable binding sites: opportunities for G protein-coupled receptor (GPCR) medicinal chemistry. J Med Chem 60:4126–4134
Li G, Aschenbach LC, Chen J, Cassidy MP, Stevens DL, Gabra BH, Selley DE, Dewey WL, Westkaemper RB, Zhang Y (2009) Design, synthesis, and biological evaluation of 6α-and 6β-N-heterocyclic substituted naltrexamine derivatives as μ opioid receptor selective antagonists. J Med Chem 52:1416–1427
Zaidi SA, Arnatt CK, He H, Selley DE, Mosier PD, Kellogg GE, Zhang Y (2013) Binding mode characterization of 6alpha- and 6beta-N-heterocyclic substituted naltrexamine derivatives via docking in opioid receptor crystal structures and site-directed mutagenesis studies: application of the 'message-address' concept in development of mu opioid receptor selective antagonists. Bioorg Med Chem 21:6405–6413
Mitra P, Venitz J, Yuan Y, Zhang Y, Gerk PM (2011) Preclinical disposition (in vitro) of novel μ-opioid receptor selective antagonists. Drug Metab Dispos 39:1589–1596
Altarifi AA, Yuan Y, Zhang Y, Selley DE, Negus SS (2015) Effects of the novel, selective and low-efficacy mu opioid receptor ligand NAQ on intracranial self-stimulation in rats. Psychopharmacology 232:815–824
Obeng S, Yuan Y, Jali A, Selley DE, Zhang Y (2018) In vitro and in vivo functional profile characterization of 17-cyclopropylmethyl-3, 14β-dihydroxy-4, 5α-epoxy-6α-(isoquinoline-3-carboxamido) morphinan (NAQ) as a low efficacy mu opioid receptor modulator. Eur J Pharmacol 827:32–40
Yuan Y, Li G, He H, Stevens DL, Kozak P, Scoggins KL, Mitra P, Gerk PM, Selley DE, Dewey WL, Zhang Y (2011) Characterization of 6alpha- and 6beta-N-heterocyclic substituted naltrexamine derivatives as novel leads to development of mu opioid receptor selective antagonists. ACS Chem Neurosci 2:346–351
Yuan Y, Zaidi SA, Stevens DL, Scoggins KL, Mosier PD, Kellogg GE, Dewey WL, Selley DE, Zhang Y (2015) Design, syntheses, and pharmacological characterization of 17-cyclopropylmethyl-3,14beta-dihydroxy-4,5alpha-epoxy-6alpha-(isoquinoline-3'-ca rboxamido)morphinan analogues as opioid receptor ligands. Bioorg Med Chem 23:1701–1715
Yuan Y, Elbegdorj O, Beletskaya IO, Selley DE, Zhang Y (2013) Structure activity relationship studies of 17-cyclopropylmethyl-3,14beta-dihydroxy-4,5alpha-epoxy-6alpha-(isoquinoline-3'-ca rboxamido)morphinan (NAQ) analogues as potent opioid receptor ligands: preliminary results on the role of electronic characteristics for affinity and function. Bioorg Med Chem Lett 23:5045–5048
Jones A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S (2012) Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature 485:321–326
Clark M, Cramer RD III, Van Opdenbosch N (1989) Validation of the general purpose tripos 5.2 force field. J Comput Chem 10:982–1012
Jones G, Willett P, Glen RC (1995) Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol 245:43–53
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748
Korb O, Stutzle T, Exner TE (2009) Empirical scoring functions for advanced protein-ligand docking with PLANTS. J Chem Inf Model 49:84–96
Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29:1859–1865
Wu EL, Cheng X, Jo S, Rui H, Song KC, Davila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, Klauda JB, Im W (2014) CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J Comput Chem 35:1997–2004
Jo S, Lim JB, Klauda JB, Im W (2009) CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys J 97:50–58
Jo S, Kim T, Im W (2007) Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE 2:e880
Case D, Berryman J, Betz R, Cerutti D, Cheatham Iii T, Darden T, Duke R, Giese T, Gohlke H, Goetz A (2015) AMBER 2015. University of California, San Francisco
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174
Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 11:3696–3713
Dickson CJ, Madej BD, Skjevik AA, Betz RM, Teigen K, Gould IR, Walker RC (2014) Lipid14: the amber lipid force field. J Chem Theory Comput 10(2):865–879
Le Grand S, Götz AW, Walker RC (2013) SPFP: Speed without compromise—a mixed precision model for GPU accelerated molecular dynamics simulations. Comput Phys Commun 184:374–380
Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC (2013) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput 9:3878–3888
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897
Weiser J, Shenkin PS, Still WC (1999) Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J Comput Chem 20:217–230
Geng L, Gao J, Cui W, Tang Y, Ji M, Chen B (2012) Computational insights into the selectivity mechanism of APP-IP over matrix metalloproteinases. J Comput Aided Mol Des 26:1327–1342
Wang H, Guo C, Chen B-Z, Ji M (2015) Computational study on the drug resistance mechanism of HCV NS5B RNA-dependent RNA polymerase mutants V494I, V494A, M426A, and M423T to Filibuvir. Antiviral Res 113:79–92
Wang H, Kellogg GE, Xu P, Zhang Y (2018) Exploring the binding mechanisms of diaminopimelic acid analogs to meso-diaminopimelate dehydrogenase by molecular modeling. J Mol Graph Modell 83:100–111
Gohlke H, Case DA (2004) Converging free energy estimates: MM-PB (GB) SA studies on the protein–protein complex Ras-Raf. J Comput Chem 25:238–250
Case DA, Cheatham TE III, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688
Wang H, Zaidi SA, Zhang Y (2017) Binding mode analyses of NAP derivatives as mu opioid receptor selective ligands through docking studies and molecular dynamics simulation. Biorg Med Chem 25:2463–2471
Gohlke H, Kiel C, Case DA (2003) Insights into protein–protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras–RalGDS complexes. J Mol Biol 330:891–913
Hou T, McLaughlin W, Lu B, Chen K, Wang W (2006) Prediction of binding affinities between the human amphiphysin-1 SH3 domain and its peptide ligands using homology modeling, molecular dynamics and molecular field analysis. J Proteome Res 5:32–43
Zoete V, Irving M, Michielin O (2010) MM–GBSA binding free energy decomposition and T cell receptor engineering. J Mol Recognit 23:142–152
Fedorov DG, Kitaura K (2018) Pair interaction energy decomposition analysis for density functional theory and density-functional tight-binding with an evaluation of energy fluctuations in molecular dynamics. J Phys Chem A 122:1781–1795
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652
Lee C, Yang W, Parr RG (1988) Development of the Colle–Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37:785–789
Vosko SH, Wilk L, Nusair M (1980) Accurate spin-dependent electron liquid correlation energies for local spin density calculations: a critical analysis. Can J Phys 58:1200–1211
Frisch M, Trucks G, Schlegel H, Scuseria Gw, Robb M, Cheeseman J, Montgomery J, Vreven T, Kudin K, Burant J (2008) Gaussian 03, revision C. 02
Utesch T, de Miguelcatalina A, Schattenberg C, Paege N, Schmieder P, Krause E, Miao Y, McCammon JA, Meyer V, Jung S, Mroginski MA (2018) A computational modeling approach predicts interaction of the antifungal protein AFP from Aspergillus giganteus with fungal membranes via its γ-core motif. mSphere 3:e0037-18
Feinberg EN, Farimani AB, Uprety R, Hunkele A, Pasternak GW, Majumdar S, Pande VS (2018) Machine learning harnesses molecular dynamics to discover new mu opioid chemotypes. arXiv 1803.04479.
Latorraca NR, Wang JK, Bauer B, Townshend RJ, Hollingsworth SA, Olivieri JE, Xu HE, Sommer ME, Dror RO (2018) Molecular mechanism of GPCR-mediated arrestin activation. Nature 557:452–456
Miao Y, Huang YM, Walker RC, McCammon JA, Chang CE (2018) Ligand binding pathways and conformational transitions of the HIV protease. Biochemistry 57:1533–1541
Obeng S, Jali A, Zheng Y, Wang H, Schwienteck KL, Chen C, Stevens DL, Akbarali HI, Dewey WL, Banks ML, Liu-Chen LY, Selley DE, Zhang Y (2019) Characterization of 17-cyclopropylmethyl-3, 14β-dihydroxy-4, 5α-epoxy-6α-(indole-7-carboxamido) morphinan (NAN) as a novel opioid receptor modulator for opioid use disorder treatment. ACS Chem Neurosci 10:2518–2532
Obeng S, Wang H, Jali A, Stevens DL, Akbarali HI, Dewey WL, Selley DE, Zhang Y (2018) Structure–activity relationship studies of 6α-and 6β-indolylacetamidonaltrexamine derivatives as bitopic mu opioid receptor modulators and elaboration of the “message-address concept” to comprehend their functional conversion. ACS Chem Neurosci 10:1075–1090
Acknowledgements
The work was partially supported by PHS Grant DA024022 (Y. Z.), DA050311 (Y. Z.), and the VCU Center for High-Performance Computing (CHiPC). The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Drug Abuse or the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors do not have any conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, H., Reinecke, B.A. & Zhang, Y. Computational insights into the molecular mechanisms of differentiated allosteric modulation at the mu opioid receptor by structurally similar bitopic modulators. J Comput Aided Mol Des 34, 879–895 (2020). https://doi.org/10.1007/s10822-020-00309-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-020-00309-x