Abstract
A20 is an anti-inflammatory protein that is strongly linked to human disease. Here, we find that mice expressing three distinct targeted mutations of A20’s zinc finger 7 (ZF7) ubiquitin-binding motif uniformly developed digit arthritis with features common to psoriatic arthritis, while mice expressing point mutations in A20’s OTU or ZF4 motifs did not exhibit this phenotype. Arthritis in A20ZF7 mice required T cells and MyD88, was exquisitely sensitive to tumor necrosis factor and interleukin-17A, and persisted in germ-free conditions. A20ZF7 cells exhibited prolonged IκB kinase activity that drove exaggerated transcription of late-phase nuclear factor-κB response genes in vitro and in prediseased mouse paws in vivo. In addition, mice expressing double-mutant A20 proteins in A20’s ZF4 and ZF7 motifs died perinatally with multi-organ inflammation. Therefore, A20’s ZF4 and ZF7 motifs synergistically prevent inflammatory disease in a non-catalytic manner.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Malynn, B. A. & Ma, A. A20: a multifunctional tool for regulating immunity and preventing disease. Cell Immunol. 340, 103914 (2019).
Catrysse, L., Vereecke, L., Beyaert, R. & van Loo, G. A20 in inflammation and autoimmunity. Trends Immunol. 35, 22–31 (2014).
Schuijs, M. J. et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 349, 1106–1110 (2015).
Aki, A., Nagasaki, M., Malynn, B. A., Ma, A. & Kagari, T. Hypomorphic A20 expression confers susceptibility to psoriasis. PLoS One 12, e0180481 (2017).
Zhou, Q. et al. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat. Genet. 48, 67–73 (2016).
Aeschlimann, F. A. et al. A20 haploinsufficiency (HA20): clinical phenotypes and disease course of patients with a newly recognised NF-κB-mediated autoinflammatory disease. Ann. Rheum. Dis. 77, 728–735 (2018).
Berteau, F. et al. Autosomic dominant familial Behçet disease and haploinsufficiency A20: a review of the literature. Autoimmun. Rev. 17, 809–815 (2018).
Kadowaki, T. et al. Haploinsufficiency of A20 causes autoinflammatory and autoimmune disorders. J. Allergy Clin. Immunol. 141, 1485–1488.e11 (2018).
Lee, E. G. et al. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science 289, 2350–2354 (2000).
Boone, D. L. et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 5, 1052–1060 (2004).
Lin, S.-C. et al. Molecular basis for the unique deubiquitinating activity of the NF-κB inhibitor A20. J. Mol. Biol. 376, 526–540 (2008).
Wertz, I. E. et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, 694–699 (2004).
Bosanac, I. et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol. Cell 40, 548–557 (2010).
Tokunaga, F. et al. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 31, 3856–3870 (2012).
Verhelst, K. et al. A20 inhibits LUBAC-mediated NF-κB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 31, 3845–3855 (2012).
Shembade, N., Ma, A. & Harhaj, E. W. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 327, 1135–1139 (2010).
Skaug, B. et al. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol. Cell 44, 559–571 (2011).
Lu, T. T. et al. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity 38, 896–905 (2013).
Draber, P. et al. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 13, 2258–2272 (2015).
Wertz, I. E. et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 528, 370–375 (2015).
De, A., Dainichi, T., Rathinam, C. V. & Ghosh, S. The deubiquitinase activity of A20 is dispensable for NF-κB signaling. EMBO Rep. 15, 775–783 (2014).
Turer, E. E. et al. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J. Exp. Med. 205, 451–464 (2008).
Kool, M. et al. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity 35, 82–96 (2011).
Hammer, G. E. et al. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat. Immunol. 12, 1184–1193 (2011).
Duong, B. H. et al. A20 restricts ubiquitination of pro-interleukin-1β protein complexes and suppresses NLRP3 inflammasome activity. Immunity 42, 55–67 (2015).
Vande Walle, L. et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature 512, 69–73 (2014).
Blauvelt, A. & Chiricozzi, A. The immunologic role of IL-17 in psoriasis and psoriatic arthritis pathogenesis. Clin. Rev. Allergy Immunol. 55, 379–390 (2018).
Robert, M. & Miossec, P. IL-17 in rheumatoid arthritis and precision medicine: from synovitis expression to circulating bioactive levels. Front. Med. 5, 364 (2018).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Wu, H.-J. et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010).
Scher, J. U. et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2, e01202 (2013).
Elliott, M. J. et al. Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor α(cA2) versus placebo in rheumatoid arthritis. Lancet 344, 1105–1110 (1994).
Ritchlin, C. T., Colbert, R. A. & Gladman, D. D. Psoriatic arthritis. N. Engl. J. Med. 376, 957–970 (2017).
Matmati, M. et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat. Genet. 43, 908–912 (2011).
Hoffmann, A., Levchenko, A., Scott, M. L. & Baltimore, D. The IκB–NF-κB signaling module: temporal control and selective gene activation. Science 298, 1241–1245 (2002).
Hao, S. & Baltimore, D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 10, 281–288 (2009).
McInnes, I. B. & Schett, G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat. Rev. Immunol. 7, 429–442 (2007).
Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942 (2019).
Sims, J. J. & Cohen, R. E. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of Rap80. Mol. Cell 33, 775–783 (2009).
Sato, Y. et al. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28, 2461–2468 (2009).
Polykratis, A. et al. A20 prevents inflammasome-dependent arthritis by inhibiting macrophage necroptosis through its ZnF7 ubiquitin-binding domain. Nat. Cell Biol. 21, 731–742 (2019).
Veale, D. J. & Fearon, U. The pathogenesis of psoriatic arthritis. Lancet 391, 2273–2284 (2018).
Jacques, P. et al. Proof of concept: enthesitis and new bone formation in spondyloarthritis are driven by mechanical strain and stromal cells. Ann. Rheum. Dis. 73, 437–445 (2014).
Armaka, M. et al. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. J. Exp. Med. 205, 331–337 (2008).
Plenge, R. M. et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat. Genet. 39, 1477–1482 (2007).
Zilberman-Rudenko, J. et al. Recruitment of A20 by the C-terminal domain of NEMO suppresses NF-κB activation and autoinflammatory disease. Proc. Natl Acad. Sci. USA 113, 1612–1617 (2016).
Emmerich, C. H. et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl Acad. Sci. USA 110, 15247–15252 (2013).
Tavares, R. M. et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity 33, 181–191 (2010).
Onizawa, M. et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat. Immunol. 16, 618–627 (2015).
Acknowledgements
We thank J. Ashouri Sinha, J. Zikherman, H.-E. Liang and J. Zhang for helpful discussions and advice and N. Szeto for the micro-CT reconstructions. This work was supported by NIH R01 AI135198 and R01 AI117908 (to A.M.), AR070155 (to M.C.N.) and a Dermatology Foundation Fellowship (to B.R.). M.C.N. is also supported by the Russell/Engleman Arthritis Center at UCSF and the Department of Veterans Affairs Health Care System. B.R. is also supported by the Department of Veterans Affairs Health Care System. Micro-CT analyses were performed by the UCSF Core Center for Musculoskeletal Biology and Medicine, supported by P30AR066262 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases. The FACS analyses were supported by UCSF Liver Center grant P30DK026743 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Author information
Authors and Affiliations
Contributions
B.R. and M.I.W. helped to conceptualize the project, conducted experiments, analyzed data and helped to write the manuscript. F.S.K. and X.S. performed experiments and analyzed data. M.C.N. analyzed and interpreted gross and histological data. R.A., P.T., P.A. and M.P. helped to conduct experiments. J.A.T. conducted experiments with germ-free mice under the supervision of P.J.T. B.A.M. and A.M. conceptualized the overall project, acquired funding for the project, generated the gene-targeted mice, supervised experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
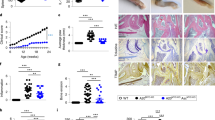
Extended Data Fig. 1 Arthritis in A20ZF7/ZF7 mice.
a, Quantitation of live born pups at 12 days of age. b, Longitudinal quantitation of digital disease of A20ZF7/ZF7 (ZF7) mice by numbers of digits with dactylitis (distal digit swelling) and nail loss in male and female mice at the indicated ages (from 4-12 weeks of age, Mean ± SD for independent mice, n = 10 for each genotype). c, Gross photos (left) and H&E histology (right) of forepaws of A20ZF7-ΔFGN/ZF7-ΔFGN and A20ZF7-FG/ZF7-FG mice at 3 months of age. Scale bars = 1 mm. Representative of three independent mice.
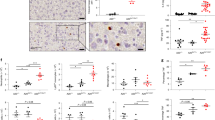
Extended Data Fig. 2 Mild systemic inflammation in A20ZF7/ZF7 mice.
Representative histology (H&E) from Kidney (a), Liver (b), Colon (c), and Lung (d) of wild-type and ZF7 mice at 3months of age. Scale bars are 100 μm for Kidney and Liver, 200 μm for Colon, and 1 mm for Lung. Representative of three independent mice.
Extended Data Fig. 3 WT and ZF7 T cell signaling and T cell profiling.
a, Flow cytometry analysis of TCRβ+ and TCRδ+ T cells in lymph nodes (axillary, brachial and inguinal combined, left) and total quantitation of TCRδ+ cells in lymph nodes (right). b, (left) Representative flow cytometry analysis of naive CD4+ T cells after separation from spleen and lymph nodes (EasySep Mouse Naive CD4+ T cell Isolation Kit). (right) Immunoblot analysis of TCR signaling from purified naive CD4+ T cells shown at left. Purified naive CD4+ T cells were stimulated with anti-CD3 mAb (145-2C11; Tonbo) followed by crosslinking with goat anti-hamster IgG (Jackson Immuno) for the indicated times at 37 °C with gentle shaking. Data are representative of three independent experiments.
Extended Data Fig. 4 Myeloid activation status in wild-type and A20ZF7/ZF7 mice.
Representative flow cytometry analysis of I-A/E cell surface expression on CD11b+ Gr1+ a, and CD11chi b, cells in the spleen of 3-month old WT and ZF7 mice. Cells were gated on lineage negative population (TCRβ, CD19). Data are representative of 5 biological replicates per genotype.
Extended Data Fig. 5 Quantitation of T cell subsets in 3-month old wild-type and A20ZF7/ZF7 mice.
a, Flow cytometric analysis of CD25 vs FoxP3 expression on CD4+ T cells. b, Flow cytometric analysis of intracellular IFNγ on ex vivo CD4+ T cells stimulated with PMA and ionomycin. Data are representative of two independent experiments.
Extended Data Fig. 6 Expression of early NF-κB -induced genes.
(a) Quantitative qPCR analyses of indicated early NF-κB-induced genes in mouse embryonic fibroblasts (MEFs) transiently stimulated with TNF for 15 minutes. Mean ± SD for technical replicates. Results are representative from three independent experiments. (b) Quantitative PCR analyses of indicated genes in paws from pre-diseased 3-week-old mice of the indicated genotypes. Data are representative of paws from multiple independent mice (WT n = 5, ZF7, ZF4, OTU n = 6). * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. One-way ANOVA with Tukey’s multiple comparison test.
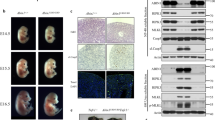
Extended Data Fig. 7 Liver inflammation in A20ZF4ZF7/ZF4ZF7 mice.
Gross (a) and hepatic H&E histology (b) of A20ZF4ZF7/ZF4ZF7 and WT littermate mice. Scale bar = 100 μM. Representative of three independent mice.
Extended Data Fig. 8 Gating strategy for flow cytometric analysis.
Representative gating strategy for flow cytometric analysis of intracellular IL-17A staining is shown.
Supplementary information
Source data
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Razani, B., Whang, M.I., Kim, F.S. et al. Non-catalytic ubiquitin binding by A20 prevents psoriatic arthritis–like disease and inflammation. Nat Immunol 21, 422–433 (2020). https://doi.org/10.1038/s41590-020-0634-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0634-4
This article is cited by
-
The Complexity of Being A20: From Biological Functions to Genetic Associations
Journal of Clinical Immunology (2024)
-
Protein citrullination and NET formation do not contribute to the pathology of A20/TNFAIP3 mutant mice
Scientific Reports (2023)
-
Death by TNF: a road to inflammation
Nature Reviews Immunology (2023)
-
A20 and ABIN-1 cooperate in balancing CBM complex-triggered NF-κB signaling in activated T cells
Cellular and Molecular Life Sciences (2022)
-
Ankylosing spondylitis: an autoimmune or autoinflammatory disease?
Nature Reviews Rheumatology (2021)