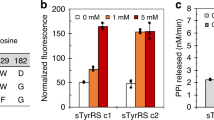
Abstract
Tyrosine sulfation is an important post-translational modification found in higher eukaryotes. Here we report an engineered tyrosyl-tRNA synthetase/tRNA pair that co-translationally incorporates O-sulfotyrosine in response to UAG codons in Escherichia coli and mammalian cells. This platform enables recombinant expression of eukaryotic proteins homogeneously sulfated at chosen sites, which was demonstrated by expressing human heparin cofactor II in mammalian cells in different states of sulfation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Data associated with this work are available upon request from the corresponding author.
References
Moore, K. L. Protein tyrosine sulfation: a critical posttranslation modification in plants and animals. Proc. Natl Acad. Sci. USA 106, 14741–14742 (2009).
Seibert, C. & Sakmar, T. P. Toward a framework for sulfoproteomics: synthesis and characterization of sulfotyrosine-containing peptides. Biopolymers 90, 459–477 (2008).
Stone, M. J., Chuang, S., Hou, X., Shoham, M. & Zhu, J. Z. Tyrosine sulfation: an increasingly recognised post-translational modification of secreted proteins. New Biotechnol. 25, 299–317 (2009).
Yang, Y. S. et al. Tyrosine sulfation as a protein post-translational modification. Molecules (Basel, Switzerland) 20, 2138–2164 (2015).
Farzan, M. et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96, 667–676 (1999).
Huang, C.-c et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl Acad. Sci. USA 101, 2706–2711 (2004).
Li, X., Hitomi, J. & Liu, C. C. Characterization of a sulfated anti-HIV antibody using an expanded genetic code. Biochemistry 57, 2903–2907 (2018).
Stone, M. J. & Payne, R. J. Homogeneous sulfopeptides and sulfoproteins: synthetic approaches and applications to characterize the effects of tyrosine sulfation on biochemical function. Acc. Chem. Res. 48, 2251–2261 (2015).
Thompson, R. E. et al. Tyrosine sulfation modulates activity of tick-derived thrombin inhibitors. Nat. Chem. 9, 909–917 (2017).
Mikkelsen, J., Thomsen, J. & Ezban, M. Heterogeneity in the tyrosine sulfation Chinese hamster ovary cell produced recombinant FVIII. Biochemistry 30, 1533–1537 (1991).
Chin, J. W. Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017).
Italia, J. S. et al. Expanding the genetic code of mammalian cells. Biochem. Soc. Trans. 45, 555–562 (2017).
Young, D. D. & Schultz, P. G. Playing with the molecules of life. ACS Chem. Biol. 13, 854–870 (2018).
Liu, C. C., Brustad, E., Liu, W. & Schultz, P. G. Crystal structure of a biosynthetic sulfo-hirudin complexed to thrombin. J. Am. Chem. Soc. 129, 10648–10649 (2007).
Liu, C. C. & Schultz, P. G. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat. Biotechnol. 24, 1436–1440 (2006).
Watson, E. E. et al. Mosquito-derived anophelin sulfoproteins are potent antithrombotics. ACS Central Sci. 4, 468–476 (2018).
Italia, J. S., Latour, C., Wrobel, C. J. & Chatterjee, A. Resurrecting the bacterial tyrosyl-tRNA synthetase/tRNA pair for expanding the genetic code of both E. coli and eukaryotes. Cell Chem. Biol. 25, 1304–1312 (2018).
Chin, J. W. et al. An expanded eukaryotic genetic code. Science 301, 964–967 (2003).
Dumas, A., Lercher, L., Spicer, C. D. & Davis, B. G. Designing logical codon reassignment—expanding the chemistry in biology. Chem. Sci. 6, 50–69 (2015).
Italia, J. S. et al. An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nat. Chem. Biol. 13, 446–450 (2017).
Tollefsen, D. M. Heparin cofactor II. Adv. Exp. Med. Biol. 425, 35–44 (1997).
Tollefsen, D. M. Heparin cofactor II modulates the response to vascular injury. Arterioscler. Thromb. Vasc. Biol. 27, 454–460 (2007).
Hortin, G., Tollefsen, D. & Strauss, A. W. Identification of two sites of sulfation of human heparin cofactor II. J. Biol. Chem. 261, 15827–15830 (1986).
Ciaccia, A. V., Monroe, D. M. & Church, F. C. Arginine 200 of heparin cofactor II promotes intramolecular interactions of the acidic domain implication for thrombin inhibition. J. Biol. Chem. 272, 14074–14079 (1997).
Mitchell, J. W. & Church, F. C. Aspartic acid residues 72 and 75 and tyrosine-sulfate 73 of heparin cofactor II promote intramolecular interactions during glycosaminoglycan binding and thrombin inhibition. J. Biol. Chem. 277, 19823–19830 (2002).
Zheng, Y., Lewis, T. L. Jr, Igo, P., Polleux, F. & Chatterjee, A. Virus-enabled optimization and delivery of the genetic machinery for efficient unnatural amino acid mutagenesis in mammalian cells and tissues. ACS Synth. Biol. 6, 13–18 (2016).
Acknowledgements
We thank D.M. Monroe III (UNC) for helpful discussions on the HCII kinetic assay. This work was supported by the National Institutes of Health (R01GM124319 and R01GM126220 to A.C.; 1R01GM118431 and 1R01GM117004 to E.W.).
Author information
Authors and Affiliations
Contributions
A.C. and J.S.I. designed the experiments. J.S.I. conducted all experiments. C.M.H. and C.L. assisted with cloning and protein expression. J.C.P. performed the MS analyses of HCII and E.W. supervised. A.C. and J.S.I. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Table 1 and Note.
Rights and permissions
About this article
Cite this article
Italia, J.S., Peeler, J.C., Hillenbrand, C.M. et al. Genetically encoded protein sulfation in mammalian cells. Nat Chem Biol 16, 379–382 (2020). https://doi.org/10.1038/s41589-020-0493-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0493-1
This article is cited by
-
Enhanced tyrosine sulfation is associated with chronic kidney disease-related atherosclerosis
BMC Biology (2023)
-
Histone tyrosine sulfation by SULT1B1 regulates H4R3me2a and gene transcription
Nature Chemical Biology (2023)
-
Unleashing the potential of noncanonical amino acid biosynthesis to create cells with precision tyrosine sulfation
Nature Communications (2022)
-
Therapeutic peptides: current applications and future directions
Signal Transduction and Targeted Therapy (2022)
-
Site-specific incorporation of citrulline into proteins in mammalian cells
Nature Communications (2021)