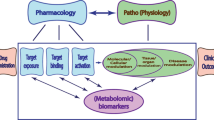
Abstract
Introduction
Pharmacogenetics and pharmacometabolomics are the common methods for personalized medicine, either genetic or metabolic biomarkers have limited predictive power for drug response.
Objectives
In order to better predict drug response, the study attempted to integrate genetic and metabolic biomarkers for drug pharmacokinetics prediction.
Methods
The study chose celecoxib as study object, the pharmacokinetic behavior of celecoxib was assessed in 48 healthy volunteers based on UPLC–MS/MS platform, and celecoxib related single nucleotide polymorphisms (SNPs) were also detected. Three mathematic models were constructed for celecoxib pharmacokinetics prediction, the first one was mainly based on celecoxib-related SNPs; the second was based on the metabolites selected from a pharmacometabolomic analysis by using GC–MS/MS method, the last model was based on the combination of the celecoxib-related SNPs and metabolites above.
Results
The result proved that the last model showed an improved prediction power, the integration model could explain 71.0% AUC variation and predict 62.3% AUC variation. To facilitate clinical application, ten potential celecoxib-related biomarkers were further screened, which could explain 68.3% and predict 54.6% AUC variation, the predicted AUC was well correlated with the measured values (r = 0.838).
Conclusion
This study provides a new route for personalized medicine, the integration of genetic and metabolic biomarkers can predict drug response with a higher accuracy.






Similar content being viewed by others
References
Aboel Dahab, A., El-Hag, D., Moutamed, G. M., Aboel Dahab, S., Abuknesha, R., & Smith, N. W. (2016). Pharmacokinetic variations in cancer patients with liver dysfunction: Applications and challenges of pharmacometabolomics. Cancer Chemotherapy and Pharmacology,78, 465–489.
Amin, A. M., Sheau-Chin, L., Azri-Mohamed-Noor, D., Sk-Abdul-Kader, M. A., Kah-Hay, Y., & Ibrahim, B. (2017). The personalization of clopidogrel antiplatelet therapy: The role of integrative pharmacogenetics and pharmacometabolomics. Cardiology Research and Practice,2017, 8062796.
Amstutz, U., Shear, N. H., Rieder, M. J., Hwang, S., Fung, V., Nakamura, H., et al. (2014). Recommendations for HLA-B*15:02 and HLA-A*31:01 genetic testing to reduce the risk of carbamazepine-induced hypersensitivity reactions. Epilepsia,55, 496–506.
Chen, R., & Snyder, M. (2013). Promise of personalized omics to precision medicine. Wiley Interdisciplinary Reviews: Systems Biology and Medicine,5, 73–82.
Chinese Pharmacopoeia Committee. (2015). Guidelines for method validation of quantitative analysis of biological samples. Pharmacopoeia of People’s Republic of China (pp. 363–368). Beijing: China Medical Science and Technology Press.
Clayton, T. A., Lindon, J. C., Cloarec, O., Antti, H., Charuel, C., Hanton, G., et al. (2006). Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature,440, 1073–1077.
Everett, J. R. (2016). From metabonomics to pharmacometabonomics: The role of metabolic profiling in personalized medicine. Frontiers in Pharmacology,7, 297.
Fer, M., Dreano, Y., Lucas, D., Corcos, L., Salaun, J. P., Berthou, F., et al. (2008). Metabolism of eicosapentaenoic and docosahexaenoic acids by recombinant human cytochromes P450. Archives of Biochemistry and Biophysics,471, 116–125.
Gong, L., Thorn, C. F., Bertagnolli, M. M., Grosser, T., Altman, R. B., & Klein, T. E. (2012). Celecoxib pathways: Pharmacokinetics and pharmacodynamics. Pharmacogenetics and Genomics,22, 310–318.
Huang, Q., Aa, J., Jia, H., Xin, X., Tao, C., Liu, L., et al. (2015). A pharmacometabonomic approach to predicting metabolic phenotypes and pharmacokinetic parameters of atorvastatin in healthy volunteers. Journal of Proteome Research,14, 3970–3981.
Kaddurah-Daouk, R., Weinshilboum, R., & Pharmacometabolomics Research Network (2015). Metabolomic signatures for drug response phenotypes: Pharmacometabolomics enables precision medicine. Clinical Pharmacology & Therapeutics,98, 71–75.
Kim, S. H., Kim, D. H., Byeon, J. Y., Kim, Y. H., Kim, D. H., Lim, H. J., et al. (2017). Effects of CYP2C9 genetic polymorphisms on the pharmacokinetics of celecoxib and its carboxylic acid metabolite. Archives of Pharmacal Research,40, 382–390.
Kimmel, S. E., French, B., Kasner, S. E., Johnson, J. A., Anderson, J. L., Gage, B. F., et al. (2013). A pharmacogenetic versus a clinical algorithm for warfarin dosing. New England Journal of Medicine,369, 2283–2293.
Liu, R., Gong, C., Tao, L., Yang, W., Zheng, X., Ma, P., et al. (2015). Influence of genetic polymorphisms on the pharmacokinetics of celecoxib and its two main metabolites in healthy Chinese subjects. European Journal of Pharmaceutical Sciences,79, 13–19.
Nicholson, J. K., Wilson, I. D., & Lindon, J. C. (2011). Pharmacometabonomics as an effector for personalized medicine. Pharmacogenomics,12, 103–111.
Oliw, E. H., Bylund, J., & Herman, C. (1996). Bisallylic hydroxylation and epoxidation of polyunsaturated fatty acids by cytochrome P450. Lipids,31, 1003–1021.
Paulson, S. K., Hribar, J. D., Liu, N. W., Hajdu, E., Bible, R. H., Jr., Piergies, A., et al. (2000). Metabolism and excretion of [(14)C]celecoxib in healthy male volunteers. Drug Metabolism and Disposition,28, 308–314.
Paulson, S. K., Kaprak, T. A., Gresk, C. J., Fast, D. M., Baratta, M. T., Burton, E. G., et al. (1999). Plasma protein binding of celecoxib in mice, rat, rabbit, dog and human. Biopharmaceutics & Drug Disposition,20, 293–299.
Phapale, P. B., Kim, S. D., Lee, H. W., Lim, M., Kale, D. D., Kim, Y. L., et al. (2010). An integrative approach for identifying a metabolic phenotype predictive of individualized pharmacokinetics of tacrolimus. Clinical Pharmacology and Therapeutics,87, 426–436.
Pirmohamed, M. (2014). Personalized pharmacogenomics: Predicting efficacy and adverse drug reactions. Annual Review of Genomics and Human Genetics,15, 349–370.
Pirmohamed, M., Burnside, G., Eriksson, N., Jorgensen, A. L., Toh, C. H., Nicholson, T., et al. (2013). A randomized trial of genotype-guided dosing of warfarin. New England Journal of Medicine,369, 2294–2303.
Prieto-Perez, R., Ochoa, D., Cabaleiro, T., Roman, M., Sanchez-Rojas, S. D., Talegon, M., et al. (2013). Evaluation of the relationship between polymorphisms in CYP2C8 and CYP2C9 and the pharmacokinetics of celecoxib. Journal of Clinical Pharmacology,53, 1261–1267.
Rodrigues, A. D. (2005). Impact of CYP2C9 genotype on pharmacokinetics: are all cyclooxygenase inhibitors the same? Drug Metabolism and Disposition,33, 1567–1575.
Shaw, K., Amstutz, U., Kim, R. B., Lesko, L. J., Turgeon, J., Michaud, V., et al. (2015). Clinical practice recommendations on genetic testing of CYP2C9 and VKORC1 variants in warfarin therapy. Therapeutic Drug Monitoring,37, 428–436.
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. A., et al. (2007). Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics,3, 211–221.
Trygg, J., Holmes, E., & Lundstedt, T. (2007). Chemometrics in metabonomics. Journal of Proteome Research,6, 469–479.
Wang, X. Q., Shen, C. L., Wang, B. N., Huang, X. H., Hu, Z. L., & Li, J. (2015). Genetic polymorphisms of CYP2C19 2 and ABCB1 C3435T affect the pharmacokinetic and pharmacodynamic responses to clopidogrel in 401 patients with acute coronary syndrome. Gene,558, 200–207.
Werner, U., Werner, D., Rau, T., Fromm, M. F., Hinz, B., & Brune, K. (2003). Celecoxib inhibits metabolism of cytochrome P450 2D6 substrate metoprolol in humans. Clinical Pharmacology and Therapeutics,74, 130–137.
Xia, J., Sinelnikov, I. V., Han, B., & Wishart, D. S. (2015). MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Research,43, W251–W257.
Xia, J., & Wishart, D. S. (2016). Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Current Protocols in Bioinformatics,55, 14101–141091.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (Nos. 81673390, 81603219, 81603196), Jiangsu Provincial Natural Science Foundation (No. BK20191322); the Open Project Program of MOE Key Laboratory of Drug Quality Control and Pharmacovigilance (No. DQCP2017MS01); “Double First-Class” University project (Nos. CPU2018GY05 and CPU2018GY34).
Author information
Authors and Affiliations
Contributions
XX supervised the experiment, manuscript writing and performed statistical data analysis; PM, JW, LT, LL supervised all experimental procedures of the clinical trial; QH supervised the experiment, analyzed the raw data and performed the metabolite identification; XQ, BZ supervised the experiment; GZ, QS supervised and designed the experiment and manuscript writing and all authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare that there are no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xing, X., Ma, P., Huang, Q. et al. Integration analysis of metabolites and single nucleotide polymorphisms improves the prediction of drug response of celecoxib. Metabolomics 16, 41 (2020). https://doi.org/10.1007/s11306-020-01659-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-020-01659-1