Abstract
Introduction
Oral cancer is a sixth commonly occurring cancer globally. The use of tobacco and alcohol consumption are being considered as the major risk factors for oral cancer. The metabolic profiling of tissue specimens for developing carcinogenic perturbations will allow better prognosis.
Objectives
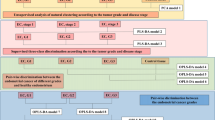
To profile and generate precise 1H HRMAS NMR spectral and quantitative statistical models of oral squamous cell carcinoma (OSCC) in tissue specimens including tumor, bed, margin and facial muscles. To apply the model in blinded prediction of malignancy among oral and neck tissues in an unknown set of patients suffering from OSCC along with neck invasion.
Methods
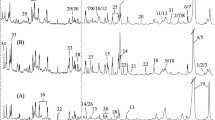
Statistical models of 1H HRMAS NMR spectral data on 180 tissues comprising tumor, margin and bed from 43 OSCC patients were performed. The combined metabolites, lipids spectral intensity and concentration-based malignancy prediction models were proposed. Further, 64 tissue specimens from twelve patients, including neck invasions, were tested for malignancy in a blinded manner.
Results
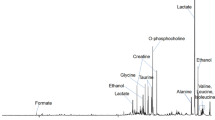
Forty-eight metabolites including lipids have been quantified in tumor and adjacent tissues. All metabolites other than lipids were found to be upregulated in malignant tissues except for ambiguous glucose. All of three prediction models have successfully identified malignancy status among blinded set of 64 tissues from 12 OSCC patients with an accuracy of above 90%.
Conclusion
The efficiency of the models in malignancy prediction based on tumor induced metabolic perturbations supported by histopathological validation may revolutionize the OSCC assessment. Further, the results may enable machine learning to trace tumor induced altered metabolic pathways for better pattern recognition. Thus, it complements the newly developed REIMS-MS iKnife real time precession during surgery.






Similar content being viewed by others
Abbreviations
- OSCC:
-
Oral squamous cell carcinoma
- HRMAS:
-
High Resolution Magic Angle Spinning
- CPMG Spectra:
-
CPMG 1H HRMAS NMR Spectra
- NOESY Spectra:
-
NOESY 1H HRMAS NMR Spectra
- OPLS-DA:
-
Orthogonal Partial Least Square Discriminant Analysis
- PLS-DA:
-
Partial Least Square Discriminant Analysis
- QUANTAS:
-
QUANTification by Artificial Signal
- CustomCSI:
-
Custom Chemical Shift Index
- SHi-PUFA:
-
Sum of Higher Polyunsaturated Fatty Acids
- MUFA:
-
Monounsaturated Fatty Acid
- SFA:
-
Saturated Fatty Acids
- TG:
-
Triglycerides
- FFA:
-
Free Fatty Acids
References
Alexandri, E., Ahmed, R., Siddiqui, H., Choudhary, M. I., Tsiafoulis, C. G., & Gerothanassis, I. P. (2017). High resolution NMR spectroscopy as a structural and analytical tool for unsaturated lipids in solution. Molecules,22(10), 1663.
Ammann, L. P., Merritt, M., Sagalowsky, A., & Nurenberg, P. (2006). Peak-finding partial least squares for the analysis of 1H NMR spectra. Journal of Chemometrics: A Journal of the Chemometrics Society,20(6–7), 231–238.
Ashizawa, K., Yoshimura, K., Johno, H., Inoue, T., Katoh, R., Funayama, S., et al. (2017). Construction of mass spectra database and diagnosis algorithm for head and neck squamous cell carcinoma. Oral Oncology,75, 111–119.
Beckwith-Hall, B. M., Brindle, J. T., Barton, R. H., Coen, M., Holmes, E., Nicholson, J. K., et al. (2002). Application of orthogonal signal correction to minimise the effects of physical and biological variation in high resolution 1H NMR spectra of biofluids. Analyst,127(10), 1283–1288.
Beger, R. D., Dunn, W., Schmidt, M. A., Gross, S. S., Kirwan, J. A., Cascante, M., et al. (2016). Metabolomics enables precision medicine: “A white paper, community perspective”. Metabolomics,12(9), 149.
Beskeen, D. W., Cram, C. M., Duffy, J., Friedrichsen, L., & Reding, E. E. (2016). Illustrated Microsoft Office 365 and Office 2016: Introductory. Boston: Cengage Learning.
Bhakoo, K. K., Williams, I. T., Williams, S. R., Gadian, D. G., & Noble, M. D. (1996). Proton nuclear magnetic resonance spectroscopy of primary cells derived from nervous tissue. Journal of Neurochemistry,66(3), 1254–1263.
Bharti, S. K., & Roy, R. (2012). Quantitative 1H NMR spectroscopy. TrAC Trends in Analytical Chemistry,35, 5–26.
Bonner, F. W., Maslen, L., Lindon, J. C., Lewis, M. R., & Nicholson, J. K. (2019). Conception, implementation and operation of large-scale metabolic phenotyping centres: phenome centres. In: The handbook of metabolic phenotyping (pp. 385–405). Amsterdam: Elsevier.
Chang, D., Weljie, A., & Newton, J. (2007). Leveraging latent information in NMR spectra for robust predictive models. In Proceedings of the Pacific symposium on biocomputing 2007 (pp. 115–126). Hackensack, NJ: World Scientific.
Chen, J.-H., Wu, Y. V., DeCarolis, P., O'Connor, R., Somberg, C. J., & Singer, S. (2008). Resolution of creatine and phosphocreatine 1H signals in isolated human skeletal muscle using HR-MAS 1H NMR. Magnetic Resonance in Medicine,59(6), 1221–1224.
Chen, X., & Yu, D. (2019). Metabolomics study of oral cancers. Metabolomics,15(2), 22.
Chenomx, N. M. R. (2015). Suite. Edmonton, AB: Chenomx Inc.
Chong, J., Soufan, O., Li, C., Caraus, I., Li, S., Bourque, G., et al. (2018). MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Research,46(W1), W486–W494.
Chong, J., Wishart, D. S., & Xia, J. (2019). Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Current Protocols in Bioinformatics,68(1), e86.
Curtius, K., Wright, N. A., & Graham, T. A. (2018). An evolutionary perspective on field cancerization. Nature Reviews Cancer,18(1), 19.
De Winter, J. C. F. (2013). Using the Student's t-test with extremely small sample sizes. Practical Assessment, Research & Evaluation,18(10), 1–12.
DeBerardinis, R. J., Lum, J. J., Hatzivassiliou, G., & Thompson, C. B. (2008). The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metabolism,7(1), 11–20.
Delikatny, E. J., Chawla, S., Leung, D. J., & Poptani, H. (2011). MR-visible lipids and the tumor microenvironment. NMR in Biomedicine,24(6), 592–611.
Farrant, R. D., Hollerton, J. C., Lynn, S. M., Provera, S., Sidebottom, P. J., & Upton, R. J. (2010). NMR quantification using an artificial signal. Magnetic Resonance in Chemistry,48(10), 753–762.
Felig, P. (1975). Amino acid metabolism in man. Annual Review of Biochemistry,44(1), 933–955.
Fernández, A., Córdova, P., Badenier, O., & Esguep, A. (2015). Epidemiological characterization of oral cancer. Literature review. Journal of Oral Research,4(2), 137–145.
Folmes, C. D. L., Nelson, T. J., & Terzic, A. (2011). Energy metabolism in nuclear reprogramming. Biomarkers in Medicine,5(6), 715–729.
Forshed, J., Idborg, H., & Jacobsson, S. P. (2007). Evaluation of different techniques for data fusion of LC/MS and 1H-NMR. Chemometrics and Intelligent Laboratory Systems,85(1), 102–109.
Gavaghan, C. L., Wilson, I. D., & Nicholson, J. K. (2002). Physiological variation in metabolic phenotyping and functional genomic studies: use of orthogonal signal correction and PLS-DA. FEBS Letters,530(1–3), 191–196.
Hong, Y. S., Coen, M., Rhode, C. M., Reily, M. D., Robertson, D. G., Holmes, E., et al. (2009). Chemical shift calibration of 1H MAS NMR liver tissue spectra exemplified using a study of glycine protection of galactosamine toxicity. Magnetic Resonance in Chemistry,47(S1), S47–S53.
Ivanisevic, J., Elias, D., Deguchi, H., Averell, P. M., Kurczy, M., Johnson, C. H., et al. (2015). Arteriovenous blood metabolomics: A readout of intra-tissue metabostasis. Scientific Reports,5, 12757.
Jafari, A., Najafi, S. H., Moradi, F., Kharazifard, M. J., & Khami, M. R. (2013). Delay in the diagnosis and treatment of oral cancer. Journal of Dentistry,14(3), 146.
Jané-Salas, E., López-López, J., Roselló-Llabrés, X., Rodríguez-Argueta, O.-F., & Chimenos-Küstner, E. (2012). Relationship between oral cancer and implants: Clinical cases and systematic literature review. Medicina Oral, Patologia Oral y Cirugia Bucal,17(1), e23.
Jiménez, B., & MacIntyre, D. (2017). NMR metabolic phenotyping in clinical studies. In J. C. Lindon, G. E. Tranter, & D. W. Koppenaal (Eds.), Encyclopedia of spectroscopy and spectrometry (3rd ed., pp. 140–145). Oxford: Academic Press.
Khanna, R., Kumar, K., & Roy, R. (2018). A case study of primary malignancy of buccal mucosa using proton HR-MAS NMR spectroscopy on tissue specimens. Journal of Oral Biology and Craniofacial Research,8(1), 68–72.
Kinross, J. M. (2019). Metabolic phenotyping in clinical practice. In The handbook of metabolic phenotyping (pp. 461–489). Amsterdam: Elsevier.
Long, G. L., & Winefordner, J. D. (1983). Limit of detection. A closer look at the IUPAC definition. Analytical Chemistry,55(7), 712A–724A.
Martens, H., & Martens, M. (2000). Modified Jack-knife estimation of parameter uncertainty in bilinear modelling by partial least squares regression (PLSR). Food Quality and Preference,11(1–2), 5–16.
Marti, H. H. (2005). Angiogenesis—a self-adapting principle in hypoxia. In M. Clauss & G. Breier (Eds.), Mechanisms of angiogenesis (pp. 163–180). Basel: Birkhäuser.
Michal, G., & Schomburg, D. (2012). Biochemical pathways: an atlas of biochemistry and molecular biology. New York: Wiley.
Mikkelsen, S. R., & Cortón, E. (2016). Bioanalytical chemistry. Hoboken: Wiley.
Moazzami, A. A., Frank, S., Gombert, A., Sus, N., Bayram, B., Rimbach, G., et al. (2015). Non-targeted 1 H-NMR-metabolomics suggest the induction of master regulators of energy metabolism in the liver of vitamin E-deficient rats. Food & Function,6(4), 1090–1097.
Nicholson, J. K., Holmes, E., Kinross, J. M., Darzi, A. W., Takats, Z., & Lindon, J. C. (2012). Metabolic phenotyping in clinical and surgical environments. Nature,491, 384.
Olszewski, W. L. (2019). Peripheral lymph: formation and immune function. Boca Raton: CRC Press.
Park, J. K., Coffey, N. J., Limoges, A., & Le, A. (2018). The heterogeneity of lipid metabolism in cancer. In A. Le (Ed.), The heterogeneity of cancer metabolism (pp. 33–55). Cham: Springer.
Paul, A., Kumar, S., Raj, A., Sonkar, A. A., Jain, S., Singhai, A., et al. (2018). Alteration in lipid composition differentiates breast cancer tissues: A 1H HRMAS NMR metabolomic study. Metabolomics,14(9), 119.
Penson, R. T. (2009). Sugar fuels cancer. Cancer,115(5), 918–921.
Polet, F., & Feron, O. (2013). Endothelial cell metabolism and tumour angiogenesis: Glucose and glutamine as essential fuels and lactate as the driving force. Journal of Internal Medicine,273(2), 156–165.
Proskuryakov, S. Y., & Gabai, V. L. (2010). Mechanisms of tumor cell necrosis. Current Pharmaceutical Design,16(1), 56–68.
Rajeev, R., Alexander, B., & Karthikeyan, S. (2015). Diagnostic methods for early detection of oral cancer: An overview. International Journal of Current Innovation Research,1(8), 197–200.
Reshef, L., Olswang, Y., Cassuto, H., Blum, B., Croniger, C. M., Kalhan, S. C., et al. (2003). Glyceroneogenesis and the triglyceride/fatty acid cycle. Journal of Biological Chemistry,278(33), 30413–30416.
Rizwan, A., & Glunde, K. (2014). Imaging of tumor metabolism: MR spectroscopy. In Functional imaging in oncology (pp. 147–180). Berlin: Springer.
Rubingh, C. M., Bijlsma, S., Derks, E. P. P. A., Bobeldijk, I., Verheij, E. R., Kochhar, S., et al. (2006). Assessing the performance of statistical validation tools for megavariate metabolomics data. Metabolomics,2(2), 53–61.
Seetharam, S. S., & Ramachandran, C. R. (1998). Fine needle aspiration cytology as a diagnostic test for oral squamous cell carcinoma. Oral Diseases,4(3), 180–186.
Shieh, A. C. (2011). Biomechanical forces shape the tumor microenvironment. Annals of Biomedical Engineering,39(5), 1379–1389.
Shield, K. D., Ferlay, J., Jemal, A., Sankaranarayanan, R., Chaturvedi, A. K., Bray, F., et al. (2017). The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA: A Cancer Journal for Clinicians,67(1), 51–64.
Slaughter, D. P., Southwick, H. W., & Smejkal, W. (1953). “Field cancerization” in oral stratified squamous epithelium. Clinical implications of multicentric origin. Cancer,6(5), 963–968.
Sreeshyla, H., Shashidar, R., & Sudhee, U. (2014). Diagnostic aids in oral precancer and cancer. Indian Journal of Multidisciplinary Dentistry,4(2), 928–934.
Srivastava, S., Roy, R., Gupta, V., Tiwari, A., Srivastava, A. N., & Sonkar, A. A. (2011). Proton HR-MAS MR spectroscopy of oral squamous cell carcinoma tissues: An ex vivo study to identify malignancy induced metabolic fingerprints. Metabolomics,7(2), 278–288.
Srivastava, S., Roy, R., Singh, S., Kumar, P., Dalela, D., Sankhwar, S. N., et al. (2010). Taurine—a possible fingerprint biomarker in non-muscle invasive bladder cancer: A pilot study by 1H NMR spectroscopy. Cancer Biomarkers,6(1), 11–20.
Sullivan, L. B., Gui, D. Y., & Vander Heiden, M. G. (2016). Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nature Reviews Cancer,16(11), 680.
Szymańska, E., Saccenti, E., Smilde, A. K., & Westerhuis, J. A. (2012). Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics,8(1), 3–16.
Tata, A., Woolman, M., Ventura, M., Bernards, N., Ganguly, M., Gribble, A., et al. (2016). Rapid detection of necrosis in breast cancer with desorption electrospray ionization mass spectrometry. Scientific Reports,6, 35374.
Ubellacker, J. M., & Morrison, S. J. (2019). Metabolic adaptation fuels lymph node metastasis. Cell Metabolism,29(4), 785–786.
Valli, A., Rodriguez, M., Moutsianas, L., Fischer, R., Fedele, V., Huang, H.-L., et al. (2015). Hypoxia induces a lipogenic cancer cell phenotype via HIF1α-dependent and-independent pathways. Oncotarget,6(4), 1920.
Van, Q. N. (2013). Current NMR strategies for biomarker discovery. In Proteomic and metabolomic approaches to biomarker discovery (pp. 87–117). Amsterdam: Elsevier.
Vander Heiden, M. G., Cantley, L. C., & Thompson, C. B. (2009). Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science,324(5930), 1029–1033.
Vitols, C., & Mercier, P. (2006). Correcting lineshapes in NMR spectra. Edmonton: CHENOMX.
Wigfield, S. M., Winter, S. C., Giatromanolaki, A., Taylor, J., Koukourakis, M. L., & Harris, A. L. (2008). PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. British Journal of Cancer,98(12), 1975.
Wishart, D. S. (2008). Quantitative metabolomics using NMR. TrAC Trends in Analytical Chemistry,27(3), 228–237.
Zhang, Q., Zhao, G., Yang, N., & Zhang, L. (2019). Fasting blood glucose levels in patients with different types of diseases. Progress in Molecular Biology and Translational Science,162, 277–292.
Zhu, Y., Chen, S., Chen, C., & Chen, L. (2017). Partial least squares (PLS) methods for abnormal detection of breast Cells. In International conference of pioneering computer scientists, engineers and educators. Berlin: Springer.
Acknowledgements
The authors are thankful to Division of SAIF, CSIR-Central Drug Research Institute, Lucknow where the 1H HRMAS NMR measurement were conducted. Mr. Anup Paul would like to thank UGC (SRF Award No. 18-12/2011(ii)EU-V) for financial assistance.
Author information
Authors and Affiliations
Contributions
AAS, AA, KG and RR designed the study. AA, KG and AAS performed the surgery and sampled the tissue specimens. SS and RR conducted the experiments. AP, and RR processed and analyzed the spectral data. NH performed the histopathology. Initial draft written by AP. AAS and RR edited and revised the paper. Project administration of the study have carried under AAS, SJ and RR. All authors carefully read and agree to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no potential conflict of interest. The disclosure of potential conflict of interest in the prescribed format has been obtained from all the author.
Ethical approval
The study was ethically approved and the work was performed in strict accordance with the guidelines of Institutional Ethical Committee of King George’s Medical University (KGMU) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The subjects were explained the study procedure and written and informed consent were obtained from them prior to the study. The authors: Anup Paul, Shatakshi Srivastava, Raja Roy, Akshay Anand, Kushagra Gaurav, Nuzzat Hussain, Sudha Jain and Abhinav A. Sonkar are aware of the ethical policy.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Paul, A., Srivastava, S., Roy, R. et al. Malignancy prediction among tissues from Oral SCC patients including neck invasions: a 1H HRMAS NMR based metabolomic study. Metabolomics 16, 38 (2020). https://doi.org/10.1007/s11306-020-01660-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-020-01660-8