Abstract
Key message
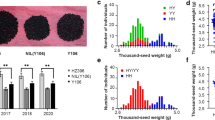
cqSW.A03-2, one of the six identified quantitative trait loci associated with thousand-seed weight in rapeseed, is mapped to a 61.6-kb region on chromosome A03 and corresponds to the candidate gene BnaA03G37960D.
Abstract
Seed weight is an important factor that determines the seed yield of oilseed rape (Brassica napus L.). To elucidate the genetic mechanism of thousand-seed weight (TSW), quantitative trait locus (QTL) mapping was conducted using a double haploid population derived from the cross between an elite line ZY50 and a pol cytoplasmic male sterility restorer line 7-5. The genetic basis of TSW was dissected into six major QTLs. One major QTL denoted as cqSW.A03-2, which explained 8.46–13.70% of the phenotypic variation, was detected across multiple environments. To uncover the genetic basis of cqSW.A03-2, a set of near-isogenic lines were developed. Based on the test of self-pollinated progenies, cqSW.A03-2 was identified as a single Mendelian factor and the ZY50 allele at cqSW.A03-2 showed a positive effect on TSW. Fine mapping delimited the cqSW.A03-2 locus into a 61.6-kb region, and 18 genes within this region were predicted. Candidate gene association analysis and expression analysis indicated that a histidine kinase gene (BnaA03G37960D) is likely to be the candidate gene for the cqSW.A03-2 locus. Our results may contribute to a better understanding of the molecular mechanism of seed weight regulation and promote the breeding program for yield improvement in rapeseed.






Similar content being viewed by others
Abbreviations
- AHK:
-
Arabidopsis histidine kinase
- BLAST:
-
Basic local alignment search tool
- CDS:
-
Coding sequences
- CIM:
-
Composite interval mapping
- cM:
-
Centimorgan
- DAP:
-
Days after pollination
- DH:
-
Double haploid
- GLM:
-
Generalized linear model
- GWAS:
-
Genome-wide association study
- InDel:
-
Insertion and deletion
- LD:
-
Linkage disequilibrium
- LOD:
-
Logarithm of the odds
- MAS:
-
Marker-assisted selection
- MBS:
-
Marker-assisted background selection
- MLM:
-
Mixed linear model
- NIL:
-
Near-isogenic line
- PCA:
-
Principal component analysis
- qRT-PCR:
-
Quantitative reverse transcription-PCR
- QTL:
-
Quantitative trait locus
- SEL:
-
Seed length
- SES:
-
Seed size
- SEW:
-
Seed width
- SL:
-
Silique length
- SN:
-
Seed number per silique
- SNP:
-
Single nucleotide polymorphism
- SSR:
-
Simple sequence repeat
- TSW:
-
Thousand-seed weight
References
Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20:2324–2326
Basunanda P, Radoev M, Ecke W, Friedt W, Becker HC, Snowdon RJ (2010) Comparative mapping of quantitative trait loci involved in heterosis for seedling and yield traits in oilseed rape (Brassica napus L.). Theor Appl Genet 120:271–281
Bazakos C, Hanemian M, Trontin C, Jimenez-Gomez JM, Loudet O (2017) New strategies and tools in quantitative genetics: how to go from the phenotype to the genotype. Annu Rev Plant Biol 68:435–455
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Brinton J, Uauy C (2019) A reductionist approach to dissecting grain weight and yield in wheat. J Integr Plant Biol 61:337–358
Cai D, Xiao Y, Yang W, Ye W, Wang B, Younas M, Wu J, Liu K (2014) Association mapping of six yieldrelated traits in rapeseed (Brassica napus L.). Theor Appl Genet 127:85–96
Cai G, Yang Q, Yi B, Fan C, Zhang C, Edwards D, Batley J, Zhou Y (2015) A bi-filtering method for processing single nucleotide polymorphism array data improves the quality of genetic map and accuracy of quantitative trait locus mapping in doubled haploid populations of polyploid Brassica napus. BMC Genom 16:409
Chalhoub B, Denoeud F, Liu S et al (2014) Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345:950–953
Chen W, Zhang Y, Liu X, Chen B, Tu J, Tingdong F (2007) Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor Appl Genet 115:849–858
Cheng X, Xu J, Xia S, Gu J, Yang Y, Fu J, Qian X, Zhang S, Wu J, Liu K (2009) Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus. Theor Appl Genet 118:1121–1131
Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X (2011) BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol 11:136
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Clarke WE, Higgins EE, Plieske J, Wieseke R, Sidebottom C, Khedikar Y, Batley J, Edwards D, Meng J, Li R, Lawley CT, Pauquet J, Laga B, Cheung W, Iniguez-Luy F, Dyrszka E, Rae S, Stich B, Snowdon RJ, Sharpe AG, Ganal MW, Parkin IA (2016) A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor Appl Genet 129:1887–1899
Cowling WA, Balazs E (2010) Prospects and challenges for genome-wide association and genomic selection in oilseed Brassica species. Genome 53:1024–1028
Dhaka N, Rout K, Yadava SK, Sodhi YS, Gupta V, Pental D, Pradhan AK (2017) Genetic dissection of seed weight by QTL analysis and detection of allelic variation in Indian and east European gene pool lines of Brassica juncea. Theor Appl Genet 130:293–307
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Elsoda M, Malosetti M, Zwaan BJ, Koornneef M, Aarts MGM (2014) Genotype × environment interaction QTL mapping in plants: lessons from Arabidopsis. Trends Plant Sci 19:390–398
Fan C, Cai G, Qin J, Li Q, Yang M, Wu J, Fu T, Liu K, Zhou Y (2010) Mapping of quantitative trait loci and development of allele-specific markers for seed weight in Brassica napus. Theor Appl Genet 121:1289–1301
Fattahi F, Fakheri BA, Solouki M, Mollers C, Rezaizad A (2018) Mapping QTL controlling agronomic traits in a doubled haploid population of winter oilseed rape (Brassica napus L.). J Genet 97:1389–1406
Fu Y, Wei D, Dong H, He Y, Cui Y, Mei J, Wan H, Li J, Snowdon R, Friedt W, Li X, Qian W (2015) Comparative quantitative trait loci for silique length and seed weight in Brassica napus. Sci Rep 5:14407
Goffinet B, Gerber S (2000) Quantitative trait loci: a meta-analysis. Genetics 155:463–473
Gupta PK, Kulwal PL, Jaiswal V (2014) Association mapping in crop plants: opportunities and challenges. Adv Genet 85:109–147
Higuchi M, Pischke MS, Mahonen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, Helariutta Y, Sussman MR, Kakimoto T (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101:8821–8826
Kearse M, Moir R, Wilson A et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kuderova A, Gallova L, Kuricova K, Nejedla E, Curdova A, Micenkova L, Plihal O, Smajs D, Spichal L, Hejatko J (2015) Identification of AHK2- and AHK3-like cytokinin receptors in Brassica napus reveals two subfamilies of AHK2 orthologues. J Exp Bot 66:339–353
Kumar J, Gupta DS, Gupta S, Dubey S, Gupta P, Kumar S (2017) Quantitative trait loci from identification to exploitation for crop improvement. Plant Cell Rep 36:1187–1213
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li F, Chen B, Xu K, Wu J, Song W, Bancroft I, Harper AL, Trick M, Liu S, Gao G, Wang N, Yan G, Qiao J, Li J, Li H, Xiao X, Zhang T, Wu X (2014a) Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res 21:355–367
Li N, Shi J, Wang X, Liu G, Wang H (2014b) A combined linkage and regional association mapping validation and fine mapping of two major pleiotropic QTLs for seed weight and silique length in rapeseed (Brassica napus L.). BMC Plant Biol 14:114
Li N, Peng W, Shi J, Wang X, Liu G, Wang H (2015) The natural variation of seed weight is mainly controlled by maternal genotype in rapeseed (Brassica napus L.). PLoS ONE 10:e0125360
Li N, Song D, Peng W, Zhan J, Shi J, Wang X, Liu G, Wang H (2019a) Maternal control of seed weight in rapeseed (Brassica napus L.): the causal link between the size of pod (mother, source) and seed (offspring, sink). Plant Biotechnol J 17:736–749
Li N, Xu R, Li Y (2019b) Molecular networks of seed size control in plants. Annu Rev Plant Biol 70:435–463
Liu Z, Liu P, Long F, Hong D, He Q, Yang G (2012) Fine mapping and candidate gene analysis of the nuclear restorer gene Rfp for pol CMS in rapeseed (Brassica napus L.). Theor Appl Genet 125:773–779
Liu J, Hua W, Hu Z, Yang H, Zhang L, Li R, Deng L, Sun X, Wang X, Wang H (2015) Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc Natl Acad Sci USA 112:E5123–E5132
Liu Z, Yang Z, Wang X, Li K, An H, Liu J, Yang G, Fu T, Yi B, Hong D (2016) A mitochondria-targeted PPR protein restores pol cytoplasmic male sterility by reducing orf224 transcript levels in oilseed rape. Mol Plant 9:1082–1084
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−△△C(T)) method. Methods 25:402–408
Lu K, Peng L, Zhang C, Lu J, Yang B, Xiao Z, Liang Y, Xu X, Qu C, Zhang K, Liu L, Zhu Q, Fu M, Yuan X, Li J (2017) Genome-wide association and transcriptome analyses reveal candidate genes underlying yield-determining traits in Brassica napus. Front Plant Sci 8:206
Luo Z, Wang M, Long Y, Huang Y, Shi L, Zhang C, Liu X, Fitt BDL, Xiang J, Mason AS, Snowdon RJ, Liu P, Meng J, Zou J (2017) Incorporating pleiotropic quantitative trait loci in dissection of complex traits: seed yield in rapeseed as an example. Theor Appl Genet 130:1569–1585
Mackay TF, Stone EA, Ayroles JF (2009) The genetics of quantitative traits: challenges and prospects. Nat Rev Genet 10:565–577
Mahonen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Tormakangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y (2006) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311:94–98
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Merk HL, Yarnes SC, Deynze AV, Tong N, Menda N, Mueller LA, Mutschler MA, Loewen SA, Myers JR, Francis DM (2012) Trait diversity and potential for selection indices based on variation among regionally adapted processing tomato germplasm. J Am Soc Hortic Sci 137:427–437
Muller B, Sheen J (2007) Arabidopsis cytokinin signaling pathway. Sci STKE 2007:cm5
Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16:1365–1377
Quijada PA, Udall JA, Lambert B, Osborn TC (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor Appl Genet 113:549–561
Raboanatahiry N, Chao H, Guo L, Gan J, Xiang J, Yan M, Zhang L, Yu L, Li M (2017) Synteny analysis of genes and distribution of loci controlling oil content and fatty acid profile based on QTL alignment map in Brassica napus. BMC Genom 18:776
Raboanatahiry N, Chao H, Dalin H, Pu S, Yan W, Yu L, Wang B, Li M (2018) QTL alignment for seed yield and yield related traits in Brassica napus. Front Plant Sci 9:1127
Radoev M, Becker HC, Ecke W (2008) Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179:1547–1558
Raman H, Raman R, Kilian A, Detering F, Long Y, Edwards D, Parkin IA, Sharpe AG, Nelson MN, Larkan N, Zou J, Meng J, Aslam MN, Batley J, Cowling WA, Lydiate D (2013) A consensus map of rapeseed (Brassica napus L.) based on diversity array technology markers: applications in genetic dissection of qualitative and quantitative traits. BMC Genom 14:277
Riefler M, Novak O, Strnad M, Schmulling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18:40–54
Shi J, Li R, Qiu D, Jiang C, Long Y, Morgan C, Bancroft I, Zhao J, Meng J (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182:851–861
Shi J, Li R, Zou J, Long Y, Meng J (2011) A dynamic and complex network regulates the heterosis of yield-correlated traits in rapeseed (Brassica napus L.). PLoS ONE 6:e21645
Shi J, Zhan J, Yang Y, Ye J, Huang S, Li R, Wang X, Liu G, Wang H (2015) Linkage and regional association analysis reveal two new tightly-linked major-QTLs for pod number and seed number per pod in rapeseed (Brassica napus L.). Sci Rep 5:14481
Shi L, Song J, Guo C, Wang B, Guan Z, Yang P, Chen X, Zhang Q, King GJ, Wang J, Liu K (2019) A CACTA-like transposable element in the upstream region of BnaA9.CYP78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J 98:524–539
Sun LJ, Wang XD, Yu KJ, Li WJ, Peng Q, Chen F, Zhang W, Fu SX, Xiong DQ, Chu P, Guan RZ, Zhang JF (2018) Mapping of QTLs controlling seed weight and seed-shape traits in Brassica napus L. using a high-density SNP map. Euphytica 214:228
Udall JA, Quijada PA, Lambert B, Osborn TC (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 2. Identification of alleles from unadapted germplasm. Theor Appl Genet 113:597–609
Wang X, Wang H, Wang J et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Wang X, Wang H, Long Y, Li D, Yin Y, Tian J, Chen L, Liu L, Zhao W, Zhao Y, Yu L, Li M (2013) Identification of QTLs associated with oil content in a high-oil Brassica napus cultivar and construction of a high-density consensus map for QTLs comparison in B. napus. PLoS ONE 8:e80569
Wang H, Xu S, Fan Y et al (2018) Beyond pathways: genetic dissection of tocopherol content in maize kernels by combining linkage and association analyses. Plant Biotechnol J 16:1464–1475
Weir B (2008) The biology of Brassica napus L. (canola). Department of Health and Aging Office of the Gene Technology Regulator, Australian Government, Canberra
Wu Y, Bhat PR, Close TJ, Lonardi S (2008) Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet 4:e1000212
Wurschum T (2012) Mapping QTL for agronomic traits in breeding populations. Theor Appl Genet 125:201–210
Xiao Y, Tong H, Yang X, Xu S, Pan Q, Qiao F, Raihan MS, Luo Y, Liu H, Zhang X, Yang N, Wang X, Deng M, Jin M, Zhao L, Luo X, Zhou Y, Li X, Liu J, Zhan W, Liu N, Wang H, Chen G, Cai Y, Xu G, Wang W, Zheng D, Yan J (2016) Genome-wide dissection of the maize ear genetic architecture using multiple populations. New Phytol 210:1095–1106
Yang P, Shu C, Chen L, Xu J, Wu J, Liu K (2012) Identification of a major QTL for silique length and seed weight in oilseed rape (Brassica napus L.). Theor Appl Genet 125:285–296
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhang L, Yang G, Liu P, Hong D, Li S, He Q (2011) Genetic and correlation analysis of silique-traits in Brassica napus L. by quantitative trait locus mapping. Theor Appl Genet 122:21–31
Zhao W, Wang X, Wang H, Tian J, Li B, Chen L, Chao H, Long Y, Xiang J, Gan J, Liang W, Li M (2016) Genome-wide identification of QTL for seed yield and yield-related traits and construction of a high-density consensus map for QTL comparison in Brassica napus. Front Plant Sci 7:17
Zhou QH, Fu DH, Mason AS, Zeng YJ, Zhao CX, Huang YJ (2014) In silico integration of quantitative trait loci for seed yield and yield-related traits in Brassica napus. Mol Breed 33:881–894
Zhou Q, Zhou C, Zheng W, Mason AS, Fan S, Wu C, Fu D, Huang Y (2017) Genome-wide SNP markers based on SLAF-Seq uncover breeding traces in rapeseed (Brassica napus L.). Front Plant Sci 8:648
Zhu M, Zhao S (2007) Candidate gene identification approach: progress and challenges. Int J Biol Sci 3:420–427
Zhu L, Zhang D, Fu T, Shen J, Wen J (2011) Analysis of yield and disease resistance traits of new winter rapeseed varieties over the past twenty years in China. Agric Sci Technol 12:842–846
Acknowledgements
We thank Dr. Liang Guo from Huazhong Agricultural University for providing phenotypic and genotypic data collected from natural population (data unpublished). This research was supported by Grants from the National 973 Project (2015CB150202).
Author information
Authors and Affiliations
Contributions
HW carried out most of the experiments, including QTL analysis, fine mapping, gene cloning and sequence analysis; MY participated phenotypic and genotypic analyses of DH population; MX participated in fine mapping; PW participated in data analysis; DH and GY designed and supervised the project. HW wrote the original draft. YL, QX, LW, DH and GY were involved in reviewing and editing of the manuscript. All authors read and contributed to the revision of manuscript.
Corresponding authors
Additional information
Communicated by Albrecht E. Melchinger.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2020_3553_MOESM9_ESM.pdf
Table S1. Primer sequences of markers used for various purposes in the study. Table S2. Descriptive statistics of TSW in parents and DH population. Table S3. Variance components for TSW in the DH population. Table S4. Summary statistics of the linkage groups. Table S5. List of all detected significant QTLs for TSW. Table S6. List of QTLs for TSW detected on A09 linkage group (PDF 163 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Yan, M., Xiong, M. et al. Genetic dissection of thousand-seed weight and fine mapping of cqSW.A03-2 via linkage and association analysis in rapeseed (Brassica napus L.). Theor Appl Genet 133, 1321–1335 (2020). https://doi.org/10.1007/s00122-020-03553-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03553-9