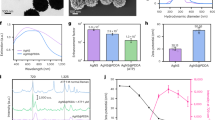
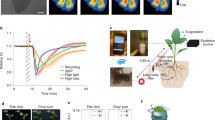
Abstract
Molecular biomarkers such as microRNAs (miRNAs) play important roles in regulating various developmental processes in plants. Understanding these pathways will help bioengineer designing organisms for efficient biomass accumulation. Current methods for RNA analysis require sample extraction and multi-step sample analysis, hindering work in field studies. Recent work in the incorporation of nanomaterials for plant bioengineering research is leading the way of an agri-tech revolution. As an example, surface-enhanced Raman scattering (SERS)–based sensors can be used to monitor RNA in vivo. However, the use of SERS in the field has been limited due to issues with observing Raman signal over complex background. To this end, shifted-excitation Raman difference spectroscopy (SERDS) offers an effective solution to extract the SERS signal from high background based on a physical approach. In this manuscript, we report the first application of SERDS on SERS sensors. We investigated this technique on SERS sensor developed for the detection of a microRNA biomarker, miR858. We tested the technique on in vitro samples and validated the technique by detecting the presence of exogenous miR858 in plants directly under ambient light in a growth chamber. The possibility of moving the detection of nucleic acid targets outside the constraints of laboratory setting enables numerous important bioengineering applications. Such applications can revolutionize biofuel production and agri-tech through the use of nanotechnology-based monitoring of plant growth, plant health, and exposure to pollution and pathogens.








Similar content being viewed by others
References
Trumbo JL, Zhang B, Stewart CN Jr. Manipulating micro RNA s for improved biomass and biofuels from plant feedstocks. Plant Biotechnology J. 2015;13(3):337–54.
Sharma D, Tiwari M, Pandey A, Bhatia C, Sharma A, Trivedi PK. MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol. 2016;171(2):944–59.
Lam PY, Tobimatsu Y, Takeda Y, Suzuki S, Yamamura M, Umezawa T, et al. Disrupting flavone synthase II alters lignin and improves biomass digestibility. Plant Physiol. 2017;174(2):972–85. https://doi.org/10.1104/pp.16.01973.
Wark AW, Lee HJ, Corn RM. Multiplexed detection methods for profiling microRNA expression in biological samples. Angew Chem Int Ed. 2008;47(4):644–52.
Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods. 2004;1(2):155.
Tran N. Fast and simple microRNA northern blots. Biochem Insights. 2009;2:BCI. S2257.
Giraldo JP, Landry MP, Faltermeier SM, McNicholas TP, Iverson NM, Boghossian AA, et al. Plant nanobionics approach to augment photosynthesis and biochemical sensing. Nat Mater. 2014;13(4):400.
Demirer GS, Zhang H, Matos JL, Goh NS, Cunningham FJ, Sung Y, et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat Nanotechnol. 2019;14(5):456–64. https://doi.org/10.1038/s41565-019-0382-5.
Lowry GV, Avellan A, Gilbertson LM. Opportunities and challenges for nanotechnology in the agri-tech revolution. Nat Nanotechnol. 2019;14(6):517.
Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277(5329):1078–81. https://doi.org/10.1126/science.277.5329.1078.
Graham D, Thompson DG, Smith WE, Faulds K. Control of enhanced Raman scattering using a DNA-based assembly process of dye-coded nanoparticles. Nat Nanotechnol. 2008;3(9):548–51. https://doi.org/10.1038/nnano.2008.189.
Kim NH, Lee SJ, Moskovits M. Aptamer-mediated surface-enhanced Raman spectroscopy intensity amplification. Nano Lett. 2010;10(10):4181–5. https://doi.org/10.1021/nl102495j.
Vohra P, Strobbia P, Ngo HT, Lee WT, Vo-Dinh T. Rapid Nanophotonics assay for head and neck cancer diagnosis. Sci Rep. 2018;8(1):11410. https://doi.org/10.1038/s41598-018-29428-0.
Holthoff EL, Stratis-Cullum DN, Hankus ME. A nanosensor for TNT detection based on molecularly imprinted polymers and surface enhanced Raman scattering. Sensors. 2011;11(3). https://doi.org/10.3390/s110302700.
Farrell ME, Strobbia P, Pellegrino PM, Cullum B. Surface regeneration and signal increase in surface-enhanced Raman scattering substrates. Appl Opt. 2017;56(3):B198–213.
Wabuyele MB, Vo-Dinh T. Detection of human immunodeficiency virus type 1 DNA sequence using plasmonics nanoprobes. Anal Chem. 2005;77(23):7810–5. https://doi.org/10.1021/ac0514671.
Wang H-N, Vo-Dinh T. Multiplex detection of breast cancer biomarkers using plasmonic molecular sentinel nanoprobes. Nanotechnology. 2009;20(6):065101.
Wang H-N, Fales AM, Vo-Dinh T. Plasmonics-based SERS nanobiosensor for homogeneous nucleic acid detection. Nanomedicine. 2015;11(4):811–4. https://doi.org/10.1016/j.nano.2014.12.012.
Wang H-N, Crawford BM, Fales AM, Bowie ML, Seewaldt VL, Vo-Dinh T. Multiplexed detection of MicroRNA biomarkers using SERS-based inverse molecular sentinel (iMS) nanoprobes. J Phys Chem C. 2016;120(37):21047–55. https://doi.org/10.1021/acs.jpcc.6b03299.
Wang H-N, Register JK, Fales AM, Gandra N, Cho EH, Boico A, et al. SERS nanosensors for in vivo detection of nucleic acid targets in a large animal model. Nano Res. 2018.
Crawford BM, Strobbia P, Wang H-N, Zentella R, Boyanov MI, Pei Z-M, et al. Plasmonic nanoprobes for in vivo multimodal sensing and bioimaging of MicroRNA within plants. ACS Appl Mater Interfaces. 2019;11(8):7743–54. https://doi.org/10.1021/acsami.8b19977.
Strobbia P, Ran Y, Crawford BM, Cupil-Garcia V, Zentella R, Wang H-N, et al. Inverse molecular sentinel-integrated fiberoptic sensor for direct and in situ detection of miRNA targets. Anal Chem. 2019;91(9):6345–52. https://doi.org/10.1021/acs.analchem.9b01350.
Wei D, Chen S, Liu Q. Review of fluorescence suppression techniques in Raman spectroscopy. Appl Spectrosc Rev. 2015;50(5):387–406.
Scaffidi JP, Gregas MK, Lauly B, Carter JC, Angel SM, Vo-Dinh T. Trace molecular detection via surface-enhanced Raman scattering and surface-enhanced resonance Raman scattering at a distance of 15 meters. Appl Spectrosc. 2010;64(5):485–92.
De Luca AC, Mazilu M, Riches A, Herrington CS, Dholakia K. Online fluorescence suppression in modulated Raman spectroscopy. Anal Chem. 2010;82(2):738–45. https://doi.org/10.1021/ac9026737.
Strobbia P, Sadler T, Odion RA, Vo-Dinh T. SERS in plain sight: a polarization modulation method for signal extraction. Anal Chem. 2019;91(5):3319–26. https://doi.org/10.1021/acs.analchem.8b04360.
Mosier-Boss P, Lieberman S, Newbery R. Fluorescence rejection in Raman spectroscopy by shifted-spectra, edge detection, and FFT filtering techniques. Appl Spectrosc. 1995;49(5):630–8.
Zhao J, Carrabba MM, Allen FS. Automated fluorescence rejection using shifted excitation Raman difference spectroscopy. Appl Spectrosc. 2002;56(7):834–45.
McCain ST, Willett RM, Brady DJ. Multi-excitation Raman spectroscopy technique for fluorescence rejection. Opt Express. 2008;16(15):10975–91.
Maiwald M, Erbert G, Klehr A, Kronfeldt HD, Schmidt H, Sumpf B, et al. Rapid shifted excitation Raman difference spectroscopy with a distributed feedback diode laser emitting at 785 nm. Appl Phys B Lasers Opt. 2006;85(4):509–12. https://doi.org/10.1007/s00340-006-2459-8.
Maiwald M, Eppich B, Fricke J, Ginolas A, Bugge F, Sumpf B, et al. Dual-wavelength Y-branch distributed Bragg reflector diode laser at 785 nanometers for shifted excitation Raman difference spectroscopy. Appl Spectrosc. 2014;68(8):838–43.
Maiwald M, Müller A, Sumpf B, Tränkle G. A portable shifted excitation Raman difference spectroscopy system: device and field demonstration. J Raman Spectrosc. 2016;47(10):1180–4. https://doi.org/10.1002/jrs.4953.
Register J, Maiwald M, Fales A, Strobbia P, Sumpf B, Vo-Dinh T. Shifted-excitation Raman difference spectroscopy for the detection of SERS-encoded gold nanostar probes. J Raman Spectrosc. 2018;49(12):1961–7. https://doi.org/10.1002/jrs.5482.
Fales AM, Yuan H, Vo-Dinh T. Development of hybrid silver-coated gold nanostars for nonaggregated surface-enhanced Raman scattering. J Phys Chem C. 2014;118(7):3708–15. https://doi.org/10.1021/jp4091393.
Sumpf B, Maiwald M, Müller A, Fricke J, Ressel P, Bugge F, et al. Comparison of two concepts for dual-wavelength DBR ridge waveguide diode lasers at 785 nm suitable for shifted excitation Raman difference spectroscopy. Appl Phys B Lasers Opt. 2015;120(2):261–9. https://doi.org/10.1007/s00340-015-6133-x.
Maiwald M, Sumpf B, Tränkle G. Rapid and adjustable shifted excitation Raman difference spectroscopy using a dual-wavelength diode laser at 785 nm. J Raman Spectrosc. 2018;49(11):1765–75. https://doi.org/10.1002/jrs.5456.
Acknowledgments
We would like to thank Tai-Ping Sun and Rodolfo Zentella from the Duke Biology Department for letting us use the growth chamber for these studies.
Funding
This material is based upon work supported by the US Department of Energy Office of Science, under Award Number DE-SC0019393. The authors received financial support by the Federal Ministry of Education and Research (BMBF) under contract 031A564C. Parts of the dual-wavelength diode laser turn-key system were developed within the project RaMBo (Raman Messsystem zur ortsspezifischen Bodenanalytik) within the consortium I4S (Intelligence for Soil) within the funding measure BonaRes (Soil as a Sustainable Resource for the Bioeconomy).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Published in the topical collection Advances in Direct Optical Detection with guest editors Antje J. Baeumner, Günter Gauglitz, and Jiri Homola.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Strobbia, P., Odion, R.A., Maiwald, M. et al. Direct SERDS sensing of molecular biomarkers in plants under field conditions. Anal Bioanal Chem 412, 3457–3466 (2020). https://doi.org/10.1007/s00216-020-02544-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02544-5