Abstract
NAD(P)-dependent steroid dehydrogenase-like (NSDHL), an essential enzyme in human cholesterol synthesis and a regulator of epidermal growth factor receptor (EGFR) trafficking pathways, has attracted interest as a therapeutic target due to its crucial relevance to cholesterol-related diseases and carcinomas. However, the development of pharmacological agents for targeting NSDHL has been hindered by the absence of the atomic details of NSDHL. In this study, we reported two X-ray crystal structures of human NSDHL, which revealed a detailed description of the coenzyme-binding site and the unique conformational change upon the binding of a coenzyme. A structure-based virtual screening and biochemical evaluation were performed and identified a novel inhibitor for NSDHL harboring suppressive activity towards EGFR. In EGFR-driven human cancer cells, treatment with the potent NSDHL inhibitor enhanced the antitumor effect of an EGFR kinase inhibitor. Overall, these findings could serve as good platforms for the development of therapeutic agents against NSDHL-related diseases.






Similar content being viewed by others
Abbreviations
- NAD:
-
Nicotinamide adenine dinucleotide
- NADP:
-
Nicotinamide adenine dinucleotide phosphate
- r.m.s.:
-
Root mean square
- SEM:
-
Standard error of mean
References
Hanukoglu I (1992) Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol 43(8):779–804. https://doi.org/10.1016/0960-0760(92)90307-5
Acimovic J, Rozman D (2013) Steroidal triterpenes of cholesterol synthesis. Molecules 18(4):4002–4017. https://doi.org/10.3390/molecules18044002
Sharpe LJ, Brown AJ (2013) Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). J Biol Chem 288(26):18707–18715. https://doi.org/10.1074/jbc.R113.479808
Gabitova L, Restifo D, Gorin A, Manocha K, Handorf E, Yang DH, Cai KQ, Klein-Szanto AJ, Cunningham D, Kratz LE, Herman GE, Golemis EA, Astsaturov I (2015) Endogenous sterol metabolites regulate growth of EGFR/KRAS-dependent Tumors via LXR. Cell Rep 12(11):1927–1938. https://doi.org/10.1016/j.celrep.2015.08.023
Ghosh S, Mukherjee S, Basu A (2015) Chandipura virus perturbs cholesterol homeostasis leading to neuronal apoptosis. J Neurochem 135(2):368–380. https://doi.org/10.1111/jnc.13208
Onyewu C, Blankenship JR, Del Poeta M, Heitman J (2003) Ergosterol biosynthesis inhibitors become fungicidal when combined with calcineurin inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob Agents Chemother 47(3):956–964
Barrett-Bee K, Dixon G (1995) Ergosterol biosynthesis inhibition: a target for antifungal agents. Acta Biochim Pol 42(4):465–479
de Souza W, Rodrigues JC (2009) Sterol biosynthesis pathway as target for anti-trypanosomatid drugs. Interdiscip Perspect Infect Dis 2009:642502. https://doi.org/10.1155/2009/642502
Roemer T, Boone C (2013) Systems-level antimicrobial drug and drug synergy discovery. Nat Chem Biol 9(4):222–231. https://doi.org/10.1038/nchembio.1205
Lim S, Oh PC, Sakuma I, Koh KK (2014) How to balance cardiorenometabolic benefits and risks of statins. Atherosclerosis 235(2):644–648. https://doi.org/10.1016/j.atherosclerosis.2014.06.001
Ramkumar S, Raghunath A, Raghunath S (2016) Statin therapy: review of safety and potential side effects. Acta Cardiol Sin 32(6):631–639
Baudry K, Swain E, Rahier A, Germann M, Batta A, Rondet S, Mandala S, Henry K, Tint GS, Edlind T, Kurtz M, Nickels JT Jr (2001) The effect of the erg26-1 mutation on the regulation of lipid metabolism in Saccharomyces cerevisiae. J Biol Chem 276(16):12702–12711. https://doi.org/10.1074/jbc.M100274200
Mo C, Valachovic M, Randall SK, Nickels JT, Bard M (2002) Protein-protein interactions among C-4 demethylation enzymes involved in yeast sterol biosynthesis. Proc Natl Acad Sci USA 99(15):9739–9744. https://doi.org/10.1073/pnas.112202799
Caldas H, Herman GE (2003) NSDHL, an enzyme involved in cholesterol biosynthesis, traffics through the Golgi and accumulates on ER membranes and on the surface of lipid droplets. Hum Mol Genet 12(22):2981–2991. https://doi.org/10.1093/hmg/ddg321
He M, Kratz LE, Michel JJ, Vallejo AN, Ferris L, Kelley RI, Hoover JJ, Jukic D, Gibson KM, Wolfe LA, Ramachandran D, Zwick ME, Vockley J (2011) Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J Clin Invest 121(3):976–984. https://doi.org/10.1172/JCI42650
Liu XY, Dangel AW, Kelley RI, Zhao W, Denny P, Botcherby M, Cattanach B, Peters J, Hunsicker PR, Mallon AM, Strivens MA, Bate R, Miller W, Rhodes M, Brown SD, Herman GE (1999) The gene mutated in bare patches and striated mice encodes a novel 3beta-hydroxysteroid dehydrogenase. Nat Genet 22(2):182–187. https://doi.org/10.1038/9700
Sukhanova A, Gorin A, Serebriiskii IG, Gabitova L, Zheng H, Restifo D, Egleston BL, Cunningham D, Bagnyukova T, Liu H, Nikonova A, Adams GP, Zhou Y, Yang DH, Mehra R, Burtness B, Cai KQ, Klein-Szanto A, Kratz LE, Kelley RI, Weiner LM, Herman GE, Golemis EA, Astsaturov I (2013) Targeting C4-demethylating genes in the cholesterol pathway sensitizes cancer cells to EGF receptor inhibitors via increased EGF receptor degradation. Cancer Discov 3(1):96–111. https://doi.org/10.1158/2159-8290.CD-12-0031
Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 276:307–326. https://doi.org/10.1016/S0076-6879(97)76066-X
Yin B, Cui DB, Zhang LJ, Jiang SQ, Machida S, Yuan YA, Wei DZ (2014) Structural insights into substrate and coenzyme preference by SDR family protein Gox2253 from Gluconobater oxydans. Proteins 82(11):2925–2935. https://doi.org/10.1002/prot.24603
Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66(Pt 2):213–221. https://doi.org/10.1107/S0907444909052925
Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53(Pt 3):240–255. https://doi.org/10.1107/S0907444996012255
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of coot. Acta Crystallogr D Biol Crystallogr 66(Pt 4):486–501. https://doi.org/10.1107/S0907444910007493
Brunger AT (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355(6359):472–475
Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66(Pt 1):12–21. https://doi.org/10.1107/S0907444909042073
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372(3):774–797. https://doi.org/10.1016/j.jmb.2007.05.022
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8(2):127–134
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118(45):11225–11236. https://doi.org/10.1021/ja9621760
Lakowicz JR, Szmacinski H, Nowaczyk K, Johnson ML (1992) Fluorescence lifetime imaging of free and protein-bound NADH. Proc Natl Acad Sci USA 89(4):1271–1275
Schrodinger LLC (2015) The PyMOL molecular graphics system. Version 1:8
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6(269):pl1. https://doi.org/10.1126/scisignal.2004088
Gyorffy B, Surowiak P, Budczies J, Lanczky A (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 8(12):e82241. https://doi.org/10.1371/journal.pone.0082241
Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, Takenoshita S, Gotoh N, Mizuno H, Sarai A, Kawano S, Yamaguchi R, Miyano S, Yokota J (2012) Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res 72(1):100–111. https://doi.org/10.1158/0008-5472.CAN-11-1403
S Rousseaux A Debernardi B Jacquiau AL Vitte A Vesin H Nagy-Mignotte D Moro-Sibilot PY Brichon S Lantuejoul P Hainaut J Laffaire A Reynies de DG Beer JF Timsit C Brambilla E Brambilla S Khochbin 2013 Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers Sci Transl Med 5(186):186ra166 Doi: 10.1126/scitranslmed.3005723
Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, Chang AC, Zhu CQ, Strumpf D, Hanash S, Shepherd FA, Ding K, Seymour L, Naoki K, Pennell N, Weir B, Verhaak R, Ladd-Acosta C, Golub T, Gruidl M, Sharma A, Szoke J, Zakowski M, Rusch V, Kris M, Viale A, Motoi N, Travis W, Conley B, Seshan VE, Meyerson M, Kuick R, Dobbin KK, Lively T, Jacobson JW, Beer DG, Director's Challenge Consortium for the Molecular Classification of Lung A (2008) Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 14(8):822–827. https://doi.org/10.1038/nm.1790
Holm L, Rosenstrom P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38 (Web Server issue): W545–549. doi: 10.1093/nar/gkq366
Orengo CA, Jones DT, Thornton JM (1994) Protein superfamilies and domain superfolds. Nature 372(6507):631–634. https://doi.org/10.1038/372631a0
Rao ST, Rossmann MG (1973) Comparison of super-secondary structures in proteins. J Mol Biol 76(2):241–256
Delvaux NA, Thoden JB, Holden HM (2018) Molecular architectures of Pen and Pal: Key enzymes required for CMP-pseudaminic acid biosynthesis in Bacillus thuringiensis. Protein Sci 27(3):738–749. https://doi.org/10.1002/pro.3368
Kavanagh KL, Jornvall H, Persson B, Oppermann U (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci 65(24):3895–3906. https://doi.org/10.1007/s00018-008-8588-y
Morimoto M, Souich C, Trinh J, McLarren KW, Boerkoel CF, Hendson G (2012) Expression profile of NSDHL in human peripheral tissues. J Mol Histol 43(1):95–106. https://doi.org/10.1007/s10735-011-9375-x
Cook PF, Cleland WW (2007) Enzyme kinetics and mechanism. Garland Science, London
Bhatia C, Oerum S, Bray J, Kavanagh KL, Shafqat N, Yue W, Oppermann U (2015) Towards a systematic analysis of human short-chain dehydrogenases/reductases (SDR): Ligand identification and structure-activity relationships. Chem Biol Interact 234:114–125. https://doi.org/10.1016/j.cbi.2014.12.013
Okada Y, Kimura T, Nakagawa T, Okamoto K, Fukuya A, Goji T, Fujimoto S, Sogabe M, Miyamoto H, Muguruma N, Tsuji Y, Okahisa T, Takayama T (2017) EGFR downregulation after anti-EGFR therapy predicts the antitumor effect in colorectal cancer. Mol Cancer Res 15(10):1445–1454. https://doi.org/10.1158/1541-7786.MCR-16-0383
Tomas A, Futter CE, Eden ER (2014) EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol 24(1):26–34. https://doi.org/10.1016/j.tcb.2013.11.002
Perinbam K, Balaram H, Guru Row TN, Gopal B (2017) Probing the influence of non-covalent contact networks identified by charge density analysis on the oxidoreductase BacC. Protein Eng Des Sel 30(3):265–272. https://doi.org/10.1093/protein/gzx006
Yamamoto K, Kusunoki M, Urabe I, Tabata S, Osaki S (2000) Crystallization and preliminary X-ray analysis of glucose dehydrogenase from Bacillus megaterium IWG3. Acta Crystallogr D Biol Crystallogr 56(Pt 11):1443–1445
McLarren KW, Severson TM, du Souich C, Stockton DW, Kratz LE, Cunningham D, Hendson G, Morin RD, Wu D, Paul JE, An J, Nelson TN, Chou A, DeBarber AE, Merkens LS, Michaud JL, Waters PJ, Yin J, McGillivray B, Demos M, Rouleau GA, Grzeschik KH, Smith R, Tarpey PS, Shears D, Schwartz CE, Gecz J, Stratton MR, Arbour L, Hurlburt J, Van Allen MI, Herman GE, Zhao Y, Moore R, Kelley RI, Jones SJ, Steiner RD, Raymond FL, Marra MA, Boerkoel CF (2010) Hypomorphic temperature-sensitive alleles of NSDHL cause CK syndrome. Am J Hum Genet 87(6):905–914. https://doi.org/10.1016/j.ajhg.2010.11.004
Cooper BF, Rudolph FB (1995) Product inhibition applications. Methods Enzymol 249:188–211
Maeting I, Schmidt G, Sahm H, Stahmann KP (2000) Role of a peroxisomal NADP-specific isocitrate dehydrogenase in the metabolism of the riboflavin overproducer Ashbya gossypii. J Mol Catalysis B 10(1):335–343. https://doi.org/10.1016/S1381-1177(00)00135-1
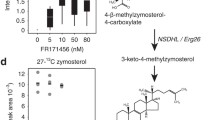
Helliwell SB, Karkare S, Bergdoll M, Rahier A, Leighton-Davis JR, Fioretto C, Aust T, Filipuzzi I, Frederiksen M, Gounarides J, Hoepfner D, Hofmann A, Imbert PE, Jeker R, Knochenmuss R, Krastel P, Margerit A, Memmert K, Miault CV, Movva NR, Muller A, Naegeli HU, Oberer L, Prindle V, Riedl R, Schuierer S, Sexton JA, Tao J, Wagner T, Yin H, Zhang J, Roggo S, Reinker S, Parker CN (2015) FR171456 is a specific inhibitor of mammalian NSDHL and yeast Erg26p. Nat Commun 6:8613. https://doi.org/10.1038/ncomms9613
Acknowledgements
We thank the beamline (BL) staff members at the Pohang Light Source, Korea (BL-5C and BL-11C), and SPring-8, Japan [beamline BL44XU under the approval of the Japan Synchrotron Radiation Research Institute (proposal No. 2017B6773)], for assistance with the X-ray diffraction experiments. All chemical libraries used in this study were kindly provided by the Korean Chemical Bank at the Korean Research Institute of Chemical Technology. This work was funded by Korea Ministry of Science, Information, Communication, Technology, and Future Planning and the National Research Foundation (NRF) of Korea Grants (NRF-2018R1A2A1A19018526 and NRF-2018R1A5A2024425 to B.-J.L.; NRF-2016R1C1B2014609, NRF-2018R1A6B4023605 and NRF-2019R1H1A1102102 to S.J.L.; and NRF-2017R1C1B2012225 to H.S.K.). This work was also supported by the 2018 BK21 Plus Project for Medicine, Dentistry, and Pharmacy and the National Cancer Center Grant of Korea (NCC-1811040; NCC-1910032; and NCC-1910023).
Author information
Authors and Affiliations
Contributions
D-GK designed and performed most experiments, analyzed data, and wrote the paper. SC initiated the project and performed crystallization of NSDHLholo and mutation studies. K-YL and S-HC helped to perform in vitro competition assay and structural analysis. H-JY collected X-ray data of crystals and solved the structure of NSDHLholo. J-YL selected compounds for in vitro competition assay from chemical library and carried out molecular docking. DK and Y-KO offered facilities and reagents for cell experiments. K-SS and C-HK guided D-GK to perform and analyze the flow cytometry experiments. YC and HHL performed SEC-MALS. Y-SJ and S-JC performed and analyzed data on the kinetic solubility of compound 9. MB and K-YJ synthesize compound 9. HSK and HJL performed and analyzed TSA experiments. SJP and J-YL offered valuable scientific feedback in the paper. J-YL also supervised experiments in her fields. SJL and B-JL oversaw all aspects of this project.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, DG., Cho, S., Lee, KY. et al. Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity. Cell. Mol. Life Sci. 78, 207–225 (2021). https://doi.org/10.1007/s00018-020-03490-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-020-03490-2