Abstract
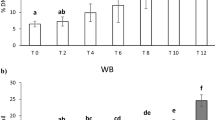
For proteome analyses, the tissue samples are mostly preserved either snap frozen or formalin-fixed, paraffin-embedded form. Use of RNAlater—a non-toxic solution primarily used to stabilize the RNA content of samples—in tissue preservation for proteome analysis recently described equally reliable with snap-frozen preservation in human tissues. Even though RNALater storage has great potential in the preservation of Peripheral Blood Mononuclear Cells (PBMC), its impact on the results of proteome analysis is poorly described at qualitative and quantitative measures. The present study investigated protein profiles of RNAlater preserved and fresh PBMCs using three extraction buffers viz. Triton X-100, RIPA and SDS. Proteins are separated in SDS-PAGE and quantified using densitometry. On an average 19.3 bands from fresh and 15.6 bands from RNAlater storage cells were obtained with a molecular weight ranging from 25 to > 250 kDa. RNAlater storage generated a fewer number and lesser quantity of low molecular weight proteins while yielded a similar or high quantity of high molecular weight protein fractions. The principal component analysis showed that Triton X-100 is inferior as compared to SDS and RIPA with respect to their protein bands and quantity yielded. While RNAlater is effective in preserving PBMC for proteome analysis, our findings warrant caution in its use in proteomics experiments especially if the target is low molecular weight proteins.









Similar content being viewed by others
References
Bennike TB, Kastaniegaard K, Padurariu S et al (2016) Comparing the proteome of snap frozen, RNAlater preserved, and formalin-fixed paraffin-embedded human tissue samples. EuPA Open Proteom 10:9–18. https://doi.org/10.1016/j.euprot.2015.10.001
Tanca A, Abbondio M, Pisanu S et al (2014) Critical comparison of sample preparation strategies for shotgun proteomic analysis of insights from liver tissue. Clin Proteom 11:28
Kruse CPS, Basu P, Luesse DR, Wyatt SE (2017) Transcriptome and proteome responses in RNAlater preserved the tissue of Arabidopsis thaliana. PLoS ONE 12:e0175943
Bennike TB, Kastaniegaard K, Padurariu S et al (2016) Proteome stability analysis of snap frozen, RNAlater preserved, and formalin-fixed paraffin-embedded human colon mucosal biopsies. Data Br 6:942–947
Saito MA, Bulygin VV, Moran DM et al (2011) Examination of microbial proteome preservation techniques applicable to autonomous environmental sample collection. Front Microbiol 2:1–10. https://doi.org/10.3389/fmicb.2011.00215
Feist P, Hummon AB (2015) Proteomic challenges: sample preparation techniques for microgram-quantity protein analysis from biological samples. Int J Mol Sci 16:3537–3563. https://doi.org/10.3390/ijms16023537
Nejadi N, Masti SM, Tavirani MR (2014) Comparison of three routine protein precipitation methods: acetone, TCA/acetone wash and TCA/acetone. J Paramed Sci 5:58–60
Buxton TB, Crockett JK, Moore WL, Rissing JP (1979) Protein precipitation by acetone for the analysis of polyethylene glycol in intestinal perfusion fluid. Gastroenterology 76:820–824
Wessel DM, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138:141–143
Arnold U, Ulbrich-Hofmann R (1999) Quantitative protein precipitation from guanidine hydrochloride-containing solutions by sodium deoxycholate/trichloroacetic acid. Anal Biochem 271:197–199
Labs Kendrick (2005) Ethanol precipitation of protein: protocol and % recovery. Kendrick Labs, Madison, pp 1–3
Rabilloud T, Luche S, Santoni V, Chevallet M (2007) Detergents and chaotropes for protein solubilization before two-dimensional electrophoresis. In: Plant proteomics. Springer, New York, pp 111–119
Abbaraju NV, Cai Y, Rees BB (2011) Protein recovery and identification from the gulf killifish, Fundulus grandis: comparing snap-frozen and RNAlaters® preserved tissues. Proteomics 11:4257–4261. https://doi.org/10.1002/pmic.201100328
Han NY, Choi W, Park JM et al (2013) Label-free quantification for discovering novel biomarkers in the diagnosis and assessment of disease activity in inflammatory bowel disease. J Dig Dis 14:166–174. https://doi.org/10.1111/1751-2980.12035
Shevchenko A, Tomas H, Havli J et al (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856
Thakur D, Rejtar T, Wang D et al (2011) Microproteomic analysis of 10,000 laser captured microdissected breast tumor cells using short-range sodium dodecyl sulfate-polyacrylamide gel electrophoresis and porous layer open tubular liquid chromatography tandem mass spectrometry. J Chromatogr A 1218:8168–8174
Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422:198
elnour A (2017) Type of stains used in detection of protein in gel. MOJ Proteom Bioinform 5:10–12. https://doi.org/10.15406/mojpb.2017.05.00163
Chevallet M, Luche S, Rabilloud T (2006) Silver staining of proteins in polyacrylamide gels. Nat Protoc 1:1852–1858. https://doi.org/10.1038/nprot.2006.288
Chevalier F (2010) Standard dyes for total protein staining in gel-based proteomic analysis. Materials 3:4784–4792. https://doi.org/10.3390/ma3104784
Mortz E, Krogh TN, Vorum H, Görg A (2001) Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteom Int Ed 1:1359–1363
Fic E, Kedracka-Krok S, Jankowska U et al (2010) Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis 31:3573–3579. https://doi.org/10.1002/elps.201000197
Zellner M, Winkler W, Hayden H et al (2005) Quantitative validation of different protein precipitation methods in proteome analysis of blood platelets. Electrophoresis 26:2481–2489
Roy VK, Senthil Kumar N, Gurusubramanian G (2012) Proteins–structure, properties and their separation by SDS-polyacrylamide gel electrophoresis. Sci Vis 12:170–181
Duncan DB (1955) Multiple range and multiple F tests. Biometrics 11:1–42
Sabullah K (2014) Comparision of staining methodsfor two dimensional electrophoresis gel resolved with Puntius javanicus liver proteome. J Biochem Microbiol Biotechnol 2(27):31
Kang D-H, Gho Y-S, Suh M-K, Kang C-H (2002) Highly sensitive and fast protein detection with coomassie brilliant blue in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Bull Korean Chem Soc 23:1511–1512
Subedi P, Schneider M, Philipp J et al (2019) Comparison of methods to isolate proteins from extracellular vesicles for mass spectrometry-based proteomic analyses. Anal Biochem 584:113390
Zhang Y, Bottinelli D, Lisacek F et al (2015) Optimization of human dendritic cell sample preparation for mass spectrometry-based proteomic studies. Anal Biochem 484:40–50. https://doi.org/10.1016/j.ab.2015.05.007
Rajeev SK, Reddy KVR (2004) Sperm membrane protein profiles of fertile and infertile men: identification and characterization of fertility-associated sperm antigen. Hum Reprod 19:234–242
Danilevich VN, Petrovskaya LE, Grishin EV (2008) A highly efficient procedure for the extraction of soluble proteins from bacterial cells with mild chaotropic solutions. Chem Eng Technol 31:904–910. https://doi.org/10.1002/ceat.200800024
Bennike TB, Carlsen TG, Ellingsen T et al (2015) Neutrophil extracellular traps in ulcerative colitis: a proteome analysis of intestinal biopsies. Inflamm Bowel Dis 21:2052–2067
Rao PK, Li Q (2009) Principal component analysis of proteome dynamics in iron-starved mycobacterium tuberculosis. J Proteom Bioinform 2:19
Zhu Y, Mullen A, Rai D et al (2019) Assessment of RNAlater® as a potential method to preserve bovine muscle proteins compared with dry ice in a proteomic study. Foods 8:60. https://doi.org/10.3390/foods8020060
van Eijsden RGE, Stassen C, Daenen L et al (2013) A universal fixation method based on quaternary ammonium salts (RNAlater) for omics-technologies: Saccharomyces cerevisiae as a case study. Biotechnol Lett 35:891–900. https://doi.org/10.1007/s10529-013-1163-0
Barclay D, Zamora R, Torres A et al (2008) A simple, rapid, and convenient luminexTM-compatible method of tissue isolation. J Clin Lab Anal 22:278–281. https://doi.org/10.1002/jcla.20253
Funding
This research work was funded by DST-SERB, Government of India; Project Number DST_SERB_ECR/2017/000761.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
Research has been approved by the Institute Animal Ethics Committee (IAEC) of the ICAR-Central Institute for Research on Cattle, Meerut, UP, India under CPCSEA guidelines. This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Alyethodi, R.R., Karthik, S., Muniswamy, K. et al. Assessment of Protein Profiles of RNAlater Stored and Fresh PBMC Cells Using Different Protein Extraction Buffers. Protein J 39, 291–300 (2020). https://doi.org/10.1007/s10930-020-09888-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-020-09888-y