Abstract
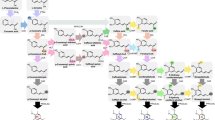
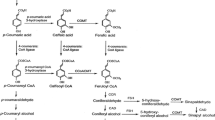
The phenylpropanoid pathway is an important route of secondary metabolism involved in the synthesis of different phenolic compounds such as phenylpropenes, anthocyanins, stilbenoids, flavonoids, and monolignols. The flux toward monolignol biosynthesis through the phenylpropanoid pathway is controlled by specific genes from at least ten families. Lignin polymer is one of the major components of the plant cell wall and is mainly responsible for recalcitrance to saccharification in ethanol production from lignocellulosic biomass. Here, we identified and characterized sugarcane candidate genes from the general phenylpropanoid and monolignol-specific metabolism through a search of the sugarcane EST databases, phylogenetic analysis, a search for conserved amino acid residues important for enzymatic function, and analysis of expression patterns during culm development in two lignin-contrasting genotypes. Of these genes, 15 were cloned and, when available, their loci were identified using the recently released sugarcane genomes from Saccharum hybrid R570 and Saccharum spontaneum cultivars. Our analysis points out that ShPAL1, ShPAL2, ShC4H4, Sh4CL1, ShHCT1, ShC3H1, ShC3H2, ShCCoAOMT1, ShCOMT1, ShF5H1, ShCCR1, ShCAD2, and ShCAD7 are strong candidates to be bona fide lignin biosynthesis genes. Together, the results provide information about the candidate genes involved in monolignol biosynthesis in sugarcane and may provide useful information for further molecular genetic studies in sugarcane.



Similar content being viewed by others
References
Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A (2011) Plant cell walls, from chemistry to biology. Ann Bot 108(1):8–9. https://doi.org/10.1093/aob/mcr128
Baltas M, Lapeyre C, Bedos-Belval F, Maturano M, Saint-Aguet P, Roussel L et al (2005) Kinetic and inhibition studies of cinnamoyl-CoA reductase 1 from Arabidopsis thaliana. Plant Physiol Biochem 43:746–753. https://doi.org/10.1016/j.plaphy.2005.06.003
Barrière Y, Argillier O (1993) Brown-midrib genes of maize: a review. Agronomie 13:865–876
Barrière Y, Riboulet C, Méchin V, Maltese S, Pichon M, Cardinal A et al (2007) Genetics and genomics of lignification in grass cell walls based on maize as model species. Genes Genomes Genom 1:133–156
Bewg WP, Coleman HD (2017) Cell wall composition and lignin biosynthetic gene expression along a developmental gradient in an Australian sugarcane cultivar. PeerJ 5:e4141. https://doi.org/10.7717/peerj.4141
Bewg WP, Poovaiah C, Lan W, Ralph J, Coleman HD (2016) RNAi down-regulation of three key lignin genes in sugarcane improves glucose release without reduction in sugar production. Biotechnol Biofuels 9:270. https://doi.org/10.1186/s13068-016-0683-y
Bi C, Chen F, Jackson L, Gill BS, Li W (2010) Expression of lignin biosynthetic genes in wheat during development and upon infection by fungal pathogens. Plant Mol Biol Rep 29:149–161. https://doi.org/10.1007/s11105-010-0219-8
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546. https://doi.org/10.1146/annurev.arplant.54.031902.134938
Bonawitz ND, Kim JI, Tobimatsu Y et al (2014) Disruption of mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509:376–380. https://doi.org/10.1038/nature13084
Bonnett GD (2014) Developmental stages (phenology). In: Moore P, Botha FC (eds) Sugarcane: physiology, biochemistry, and functional biology. Wiley, Ames, pp 35–53
Botha FC, Moore PH (2014) Biomass and bioenergy. In: Moore P, Botha FC (eds) Sugarcane: physiology, biochemistry, and functional biology. Wiley, Ames, pp 521–540
Bottcher A, Cesarino I, Santos AB, Vicentini R, Mayer JL, Vanholme R et al (2013) Lignification in sugarcane: biochemical characterization, gene discovery, and expression analysis in two genotypes contrasting for lignin content. Plant Physiol 163:1539–1557. https://doi.org/10.1104/pp.113.225250
Bout S, Vermerris W (2013) A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol Genet Genom 269:205–214. https://doi.org/10.1007/s00438-003-0824-4
Bower NI, Casu RE, Maclean DJ, Reverter A, Chapmana SC, Manners JM (2005) Transcriptional response of sugarcane roots to methyl jasmonate. Plant Sci 168:761–772. https://doi.org/10.1016/j.plantsci.2004.10.006
Brenner EA, Zein I, Chen Y, Andersen JR, Wenzel G, Ouzunova M et al (2010) Polymorphisms in O-methyltransferase genes are associated with stover cell wall digestibility in European maize (Zea mays L.). BMC Plant Biol 12:10–27. https://doi.org/10.1186/1471-2229-10-27
Brill EM, Abrahams S, Hayes CM, Jenkins CLD, Watson JM (1999) Molecular characterization and expression of a wound-inducible cDNA encoding a novel cinnamyl-alcohol dehydrogenase enzyme in lucerne. Plant Mol Biol 41:279–291
Bukh C, Nord-Larsen PH, Rasmussen SK (2012) Phylogeny and structure of the cinnamyl alcohol dehydrogenase gene family in Brachypodium distachyon. J Exp Bot 63:6223–6236. https://doi.org/10.1093/jxb/ers275
Byeon Y, Lee HY, Lee K, Back K (2014) Caffeic acid O-methyltransferase is involved in the synthesis of melatonin by methylating N-acetylserotonin in Arabidopsis. J Pineal Res 57:219–227. https://doi.org/10.1111/jpi.12160
Cantarella H, Buckeridge MS, Van Sluys MA, Souza AP, Garcia AAF, Nishiyama MY Jr et al (2012) Sugarcane. In: Kole C, Joshi CP, Shonnard DR (eds) Handbook of bioenergy crops plants. CRC Press, Boca Raton, pp 523–561
Casu RE, Grof CPL, Rae AL, McIntyre CL, Dimmock CM, Manners JM (2003) Identification of a novel sugar transporter homologue strongly expressed in maturing stem vascular tissues of sugarcane by expressed sequence tag and microarray analysis. Plant Mol Biol 52:371–386. https://doi.org/10.1023/A:1023957214644
Casu R, Dimmock CM, Chapman SC, Grof CP, McIntyre CL, Bonnett GD, Manners JM (2004) Identification of differentially expressed transcripts from maturing stem of sugarcane by in silico analysis of stem expressed sequence tags and gene expression profiling. Plant Mol Biol 54(4):503–517
Chapple CCS, Vogt T, Ellis BE, Somerville CR (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4:1413–1424
Cheavegatti-Gianotto A, de Abreu HMC, Arruda P, Bespalhok Filho JC, Burnquist WL, Creste S et al (2011) Sugarcane (Saccharum × officinarum): a reference study for the regulation of genetically modified cultivars in Brazil. Trop Plant Biol 4:62–89. https://doi.org/10.1007/s12042-011-9068-3
Chen Y, Zein I, Brenner EA, Andersen JR, Landbeck M, Ouzunova M, Lübberstedt T (2010) Polymorphisms in monolignol biosynthetic genes are associated with biomass yield and agronomic traits in European maize (Zea mays L.). BMC Plant Biol 10:12. https://doi.org/10.1186/1471-2229-10-12
Chiang VL, Funaoka M (1990) The difference between guaiacyl and guaiacyl-syringyl lignins in their response to kraft delignification. Holzforschung 44:309–313
Chiang VL, Puumala RJ, Takeuchi U, Eckert RE (1988) Comparison of softwood and hardwood kraft pulping. Tappi J 71:173–176
Chundawat SPS, Beckham GT, Himmel ME, Dale BE (2011) Deconstruction of lignocellulosic biomass to fuels and chemicals. Ann Rev Chem Biomol Eng 2:121–145. https://doi.org/10.1146/annurev-chembioeng-061010-114205
Courtial A, Soler M, Chateigner-Boutin AL, Reymond M, Mechin V, Wang H et al (2013) Breeding grasses for capacity to biofuel production or silage feeding value: an updated list of genes involved in maize secondary cell wall biosynthesis and assembly. Maydica 58:67–102
Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2014) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root-and shoot-specific genes. Plant J 38:366–379. https://doi.org/10.1111/j.1365-313X.2004.02051.x
Ehlting J, Hamberger B, Million-Rousseau R, Werck-Reichhart D (2006) Cytochromes P450 in phenolic metabolism. Phytochem Rev 5:239–270
Elkind Y, Edwards R, Mavandad M, Hedrick SA, Ribak O, Dixon RA, Lamb CJ (1990) Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc Natl Acad Sci USA 87:9057–9061
Escamilla-Treviño LL, Shen H, Uppalapati SR, Ray T, Tang Y, Hernandez T (2010) Switchgrass (Panicum virgatum) possesses a divergent family of cinnamoyl CoA reductases with distinct biochemical properties. New Phytol 185:143–155. https://doi.org/10.1111/j.1469-8137.2009.03018.x
Eudes A, Pollet B, Sibout R, Do CT, Seguin A, Lapierre C, Jouanin L (2006) Evidence for a role of AtCAD1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225:23–39. https://doi.org/10.1007/s00425-006-0326-9
Ferreira SS, Hotta CT, Poelking VG, Leite DC, Buckeridge MS, Loureiro ME et al (2016) Co-expression network analysis reveals transcription factors associated to cell wall biosynthesis in sugarcane. Plant Mol Biol 91(1–2):15–35. https://doi.org/10.1007/s11103-016-0434-2
Ferrer JL, Zubieta C, Dixon RA, Noel JP (2005) Crystal structures of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol 137:1009–1017. https://doi.org/10.1104/pp.104.048751
Fornalé S, Capellades M, Encina A, Wang K, Irar S, Lapierre C et al (2012) Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol Plant 4:817–830. https://doi.org/10.1093/mp/ssr097
Fornalé S, Rencoret J, Garcia-Calvo L, Capellades M, Encina A, Santiago R et al (2015) Cell wall modification triggere by down-regulation of coumarate 3-hydroxylase-1 in maize. Plant Sci 236:272–282. https://doi.org/10.1016/j.plantsci.2015.04.007
Fornalé S, Rencoret J, García-Calvo L, Encina A, Rigau J, Gutiérrez A et al (2017) Changes in cell wall polymers and degradability in maize mutants lacking 3′- and 5′-O-methyltransferases involved in lignin biosynthesis. Plant Cell Physiol 58(2):240–255. https://doi.org/10.1093/pcp/pcw198
Franke R, McMichael M, Meyer K, Shirley AM, Cusumano JC, Chapple C (2000) Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J 22:223–234
Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C (2002) The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 30:33–45. https://doi.org/10.1046/j.1365-313X.2002.01266.x
Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M et al (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108:3803–3808. https://doi.org/10.1073/pnas.1100310108
Garsmeur O, Droc G, Antonise R, Grimwood J, Potier B, Aitken K et al (2018) A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat Commun 9(1):2638. https://doi.org/10.1038/s41467-018-05051-5
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, Totowa, pp 571–607
Goujon T, Ferret V, Mila I, Pollet B, Ruel K, Burlat V et al (2003) Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: effects on phenotype, lignins and cell wall degradability. Planta 217:218–228. https://doi.org/10.1007/s00425-003-0987-6
Gui J, Shen J, Li L (2011) Functional characterization of evolutionarily divergent 4-coumarate: coenzyme a ligases in rice. Plant Physiol 157:574–586. https://doi.org/10.1104/pp.111.178301
Guillaumie S, San-Clemente H, Deswarte C, Martinez Y, Lapierre C, Murigneux A et al (2007) MAIZEWALL. Database and developmental gene expression profiling of cell wall biosynthesis and assembly in maize. Plant Physiol 143:339–363. https://doi.org/10.1104/pp.106.086405
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001) Down-regulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13:73–88
Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B et al (1998) Brown-midrib maize (bm1)—a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J 14(5):545–553. https://doi.org/10.1046/j.1365-313X.1998.00153.x
Hamberger B, Hahlbrock K (2004) The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc Natl Acad Sci USA 101:2209–2214. https://doi.org/10.1073/pnas.0307307101
He X, Hall MB, Gallo-Meagher M, Smith RL (2003) Improvement of forage quality by downregulation of maize O-methyltransferase. Crop Sci 43:2240–2251. https://doi.org/10.2135/cropsci2003.2240
Hirano K, Aya K, Kondo M, Okuno A, Morinaka Y, Matsuoka M (2012) OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm. Plant Cell Rep 31:91–101. https://doi.org/10.1007/s00299-011-1142-7
Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD et al (1999) Repression of lignin biosynthesis promotes cellu-lose accumulation and growth in transgenic trees. Nat Biotechnol 17:808–812
Huntley SK, Ellis D, Gilbert M, Chapple C, Mansfield SD (2003) Significant increases in pulping efficiency in C4H–F5H-transformed poplars: improved chemical savings and reduced environmental toxins. J Agric Food Chem 51:6178
Ibdah M, Zhang X, Schmidt J, Vogt T (2003) A novel Mg2+-dependent O-methyltransferase in the phenylpropanoid metabolism of Mesembryanthemum crystallinum. J Biol Chem 278:43961–43972. https://doi.org/10.1074/jbc.M304932200
Inoue K, Sewalt V, Balance G, Ni W, Stürzer K, Dixon R (1998) Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol 117:761–770
Iskandar HM, Simpson RS, Casu RE, Bonnett GD, Maclean DJ, Manners JM (2004) Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol Biol Rep 22:325–337. https://doi.org/10.1007/BF02772676
Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26:205–216. https://doi.org/10.1046/j.1365-313x.2001.01021.x
Jung HJG, Ni WT, Chapple CCS, Meyer K (1999) Impact of lignin composition on cell wall degradability in an Arabidopsis mutant. J Sci Food Agric 79:922–928
Jung JH, Fouad WM, Vermerris W, Gallo M, Altpeter F (2012) RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol J 10:1067–1076. https://doi.org/10.1111/j.1467-7652.2012.00734.x
Jung JH, Vermerris W, Gallo M, Fedenko JR, Erickson JE, Altpeter F (2013) RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field-grown sugarcane. Plant Biotechnol J 11:709–716. https://doi.org/10.1111/pbi.12061
Jung JH, Kannan B, Dermawan H, Moxley GW, Altpeter F (2016) Precision breeding for RNAi suppression of a major 4-coumarate:coenzyme A ligase gene improves cell wall saccharification from field grown sugarcane. Plant Mol Biol 92(4–5):505–517. https://doi.org/10.1007/s11103-016-0527-y
Kajita S, Hishiyama S, Tomimura Y, Katayama Y, Omori S (1997) Structural characterization of modified lignin in transgenic tobacco plants in which the activity of 4-coumarate:coenzyme A ligase is depressed. Plant Physiol 114(3):871–879
Kasirajan L, Hoang NV, Furtado A, Botha FC, Henry RJ (2018) Transcriptome analysis highlights key differentially expressed genes involved in cellulose and lignin biosynthesis of sugarcane genotypes varying in fiber content. Sci Rep 8(1):11612. https://doi.org/10.1038/s41598-018-30033-4
Kim SJ, Kim MR, Bedgar D, Moinuddin SGA, Cardenas CL, Davin LB, Kang CCK, Lewis NG (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA 101:1455–1460. https://doi.org/10.1073/pnas.0307987100
Koshiba T, Hirose N, Mukai M, Yamamura M, Hattori T, Suzuki S et al (2013) Characterization of 5-hydroxyconiferaldehyde O-methyltransferase in Oryza sativa. Plant Biotechnol 30:157–167. https://doi.org/10.5511/plantbiotechnology.13.0219a
Lallemand LA, Zubieta C, Lee SG, Wang Y, Acajjaoui S, Timmins J et al (2012) A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiol 160:249–260. https://doi.org/10.1104/pp.112.202051
Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, Grima-Pettenati J (2001) Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57:1187–1195. https://doi.org/10.1016/S0031-9422(01)00053-X
Leal MRLV, Walter AS, Seabra JE (2012) Sugarcane as an energy source. Biomass Convers Biorefin. https://doi.org/10.1007/s13399-012-0055-1
Lee D, Meyer K, Chapple C, Douglas CJ (1997) Antisense suppression of 4-coumarate: coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 9:1985–1998. https://doi.org/10.1105/tpc.9.11.1985
Levsh O, Chiang YC, Tung CF, Noel JP, Wang Y, Weng JK (2016) Dynamic conformational states dictate selectivity toward the native substrate in a substrate-permissive acyltransferase. Biochemistry 55:6314–6326. https://doi.org/10.1021/acs.biochem.6b00887
Li X, Chapple C (2010) Understanding lignification: challenges beyond monolignol biosynthesis. Plant Physiol 154:449–452. https://doi.org/10.1104/pp.110.162842
Li X, Yang Y, Yao J, Chen G, Li X, Zhang Q et al (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69:685–697. https://doi.org/10.1007/s11103-008-9448-8
Li X, Chen W, Zhao Y, Xiang Y, Jiang H, Zhu S, Cheng B (2013) Downregulation of caffeoyl-CoA O-methyltransferase (CCoAOMT) by RNA interference leads to reduced lignin production in maize straw. Genet Mol Biol 36(4):540–546. https://doi.org/10.1590/S1415-47572013005000039
Li J, Fan F, Wang L, Zhan Q, Wu P, Du J et al (2016a) Cloning and expression analysis of cinnamoyl-CoA reductase (CCR) genes in sorghum. PeerJ 4:e2005. https://doi.org/10.7717/peerj.2005
Li K, Wang H, Hu X, Liu Z, Wu Y, Huang C (2016b) Genome-wide association study reveals the genetic basis of stalk cell wall components in maize. PLoS One 11(8):e0158906. https://doi.org/10.1371/journal.pone.0158906
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Llerena JPP, Figueiredo R, Brito MDS, Kiyota E, Mayer JLS, Araujo P et al (2019) Deposition of lignin in four species of Saccharum. Sci Rep 9(1):5877. https://doi.org/10.1038/s41598-019-42350-3
Ma QH (2007) Characterization of a cinnamoyl-CoA reductase that is associated with stem development in wheat. J Exp Bot 58:2011–2021. https://doi.org/10.1093/jxb/erm064
Ma QH (2010) Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J Exp Bot 61(10):2735–2744. https://doi.org/10.1093/jxb/erq107
Ma D, Constabel CP (2019) MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci 24(3):275–289. https://doi.org/10.1016/j.tplants.2018.12.003
Ma QH, Luo HR (2015) Biochemical characterization of caffeoyl coenzyme A 3-O-methyltransferase from wheat. Planta 242:113–122. https://doi.org/10.1007/s00425-015-2295-3
Mangeon A, Lin WC, Springer PS (2012) Functional divergence in the Arabidopsis LOB-domain gene family. Plant Signal Behav 7(12):1544–1547. https://doi.org/10.4161/psb.22320
McCann MC, Carpita NC (2015) Biomass recalcitrance: a multi-scale, multi-factor, and conversion-specific property. J Exp Bot 66:4109–4118. https://doi.org/10.1093/jxb/erv267
Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C (1998) Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA 95:6619–6623
Naik SN, Goud VV, Rout PK, Dalai AK (2010) Production of first andsecond generation biofuels, a comprehensive review. Renew Sustain Energy Rev 14:578–597. https://doi.org/10.1016/j.rser.2009.10.003
Pallas JA, Paiva NL, Lamb C, Dixon RA (1996) Tobacco plants epigenetically suppressed in phenylalanine ammonia-lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. Plant J 10:281–293
Papini-Terzi FS, Rocha FR, Vêncio RZN, Felix JM, Branco DS, Waclawovsky AJ et al (2009) Sugarcane genes associated with sucrose content. BMC Genom 10:120. https://doi.org/10.1186/1471-2164-10-120
Park HL, Bhoo SH, Kwon M, Lee SW, Cho MH (2017) Biochemical and expression analyses of the rice cinnamoyl-CoA reductase gene family. Front Plant Sci 8:2099. https://doi.org/10.3389/fpls.2017.02099
Pichon M, Courbou I, Beckert M, Boudet AM, Grima-Pettenati J (1998) Cloning and characterization of two maize cDNAs encoding cinnamoyl-CoA reductase (CCR) and differential expression of the corresponding genes. Plant Mol Biol 38:671–676. https://doi.org/10.1023/A:1006060101866
Pincon G, Maury S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2001) Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth. Phytochemistry 57:1167–1176. https://doi.org/10.1016/S0031-9422(01)00098-X
Piquemal J, Chamayou S, Nadaud I, Beckert M, Barrière Y, Mila I et al (2002) Down-regulation of caffeic acid o-methyltransferase in maize revisited using a transgenic approach. Plant Physiol 130(4):1675–1685. https://doi.org/10.1104/pp.012237
Poelking VGC, Williams TCR, Rencoret J, Ralph J, Giordano A, Ricci-Silva ME et al (2015) Analysis of a modern hybrid and an ancient sugarcane implicates a complex interplay of factors in affecting recalcitrance to cellulosic ethanol production. PLoS One 10(8):e0134964. https://doi.org/10.1371/journal.pone.0134964
Poovaiah CR, Bewg WP, Lan W, Ralph J, Coleman HD (2016) Sugarcane transgenics expressing MYB transcription factors show improved glucose release. Biotechnol Biofuels 9:143. https://doi.org/10.1186/s13068-016-0559-1
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133:1051–1071. https://doi.org/10.1104/pp.103.026484
Rakoczy M, Femiak I, Alejska M, Figlerowicz M, Podkowinski J (2018) Sorghum CCoAOMT and CCoAOMT-like gene evolution, structure, expression and the role of conserved amino acids in protein activity. Mol Genet Genom 293:1077–1089. https://doi.org/10.1007/s00438-018-1441-6
Ralph J, Brunow G, Boerjan W (2007) Lignins. In: Rose F, Osborne K (eds) Encyclopedia of life sciences. Wiley, Chichester, pp 1–10. https://doi.org/10.1002/9780470015902.a0020104
Ralph J, Akiyama T, Kim H, Lu F, Schatz PF, Marita JM et al (2006) Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J Biol Chem 281:8843–8853. https://doi.org/10.1074/jbc.M511598200
Ramos RLB, Tovar FJ, Junqueira RM, Lino FB, Sachetto-Martins G (2001) Sugarcane expressed sequences tags (ESTs) encoding enzymes involved in lignin biosynthesis pathways. Genet Mol Biol 24:235–241. https://doi.org/10.1590/S1415-47572001000100031
Roach BT (1977) Utilization of Saccharum spontaneum in sugarcane breeding. Proc Int Soc Sugar Cane Technol 16:43–58
Rösler J, Krekel F, Amrhein N, Schmid J (1997) Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol 113:175–179
Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, Van Den Hoff MJ et al (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37(6):e45. https://doi.org/10.1093/nar/gkp045
Saathoff AJ, Hargrove MS, Haas EJ, Tobias CM, Twigg P, Sattler S, Sarath G (2012) Switchgrass PviCAD1: understanding residues important for substrate preferences and activity. Appl Biochem Biotechnol 168:1086–1100. https://doi.org/10.1007/s12010-012-9843-0
Saballos A, Vermerris W, Rivera L, Ejeta G (2008) Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench). Bioenergy Res 2:198–204. https://doi.org/10.1007/s12155-008-9025-7
Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W (2009) A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum (Sorghum bicolor (L.) Moench) identifies SbCAD2 as the brown midrib6 gene. Genetics 181:783–795. https://doi.org/10.1534/genetics.108.098996
Saballos A, Sattler SE, Sanchez E, Foster TP, Xin Z, Kang C et al (2012) Brown midrib2 (Bmr2) encodes the major 4-coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J 70:818–830. https://doi.org/10.1111/j.1365-313X.2012.04933.x
Santos AB, Bottcher A, Kiyota E, Mayer JL, Vicentini R, Brito Mdos S et al (2015) Water stress alters lignin content and related gene expression in two sugarcane genotypes. J Agric Food Chem 63(19):4708–4720. https://doi.org/10.1021/jf5061858
Sattler SE, Saathoff AJ, Haas EJ, Palmer NA, Funnell-Harris DL, Sarath G et al (2009) A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the Sorghum brown midrib6 phenotype. Plant Physiol 150:584–595. https://doi.org/10.1104/pp.109.136408
Sattler SE, Palmer NA, Saballos A, Greene AM (2012) Identification and characterization of four missense mutations in Brown midrib 12 (Bmr12), the Caffeic O-methyltranferase (COMT) of Sorghum. Bioenergy Res 5:855–865. https://doi.org/10.1007/s12155-012-9197-z
Sattler SE, Saballos A, Xin Z, Funnell-Harris DL, Vermerris W, Pedersen JF (2014) Characterization of Novel Sorghum brown midrib Mutants from an EMS-Mutagenized Population. G3 (Bethesda) 4(11):2115–2124. https://doi.org/10.1534/g3.114.014001
Sattler SA, Walker AM, Vermerris W, Sattler SE, Kang C (2017) Structural and biochemical characterization of cinnamoyl-CoA reductases. Plant Physiol 173(2):1031–1044. https://doi.org/10.1104/pp.16.01671
Shadle G, Chen F, Srinivasa Reddy MS, Jackson L, Nakashima J, Dixon RA (2007) Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68(11):1521–1529. https://doi.org/10.1016/j.phytochem.2007.03.022
Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL (2010) Towards a systems approach for lignin biosynthesis in populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol 51:144–163. https://doi.org/10.1093/pcp/pcp175
Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Seguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17:2059–2076. https://doi.org/10.1105/tpc.105.030767
Takeda Y, Koshiba T, Tobimatsu Y, Suzuki S, Murakami S, Yamamura M et al (2017) Regulation of CONIFERALDEHYDE 5-HYDROXYLASE expression to modulate cell wall lignin structure in rice. Planta 246(2):337–349. https://doi.org/10.1007/s00425-017-2692-x
Takeda Y, Suzuki S, Tobimatsu Y, Osakabe K, Osakabe Y, Ragamustari SK et al (2018a) Lignin characterization of rice CONIFERALDEHYDE 5-HYDROXYLASE loss-of-function mutants generated with the CRISPR/Cas9 system. Plant J 97:543–554. https://doi.org/10.1111/tpj.14141
Takeda Y, Tobimatsu Y, Karlen SD, Koshiba T, Suzuki S, Yamamura M et al (2018b) Downregulation of p-COUMAROYL ESTER 3-HYDROXYLASE in rice leads to altered cell wall structures and improves biomass saccharification. Plant J 95:796–811. https://doi.org/10.1111/tpj.13988
Tamasloukht B, Wong Quai Lam MSJ, Martinez Y, Tozo K, Barbier O, Jourda C et al (2011) Characterization of a cinnamoyl-CoA reductase 1 (CCR1) mutant in maize: effects on lignification, fibre development, and global gene expression. J Exp Bot 62:3837–3848. https://doi.org/10.1093/jxb/err077
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tew T, Cobill R (2008) Genetic improvement of Sugarcane (Saccharum spp.) as an energy crop. In: Vermerris W (ed) Genetic improvement of bioenergy crops SE-9. Springer, New York, pp 273–294
Tu Y, Rochfort S, Liu Z, Ran Y, Griffith M, Badenhorst P et al (2010) Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 22:3357–3373. https://doi.org/10.1105/tpc.109.072827
Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G et al (2012) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24:3506–3529. https://doi.org/10.1105/tpc.112.102574
Verma SR, Dwivedi UN (2014) Lignin genetic engineering for improvement of wood quality: applications in paper and textile industries, fodder and bioenergy production. S Afr J Bot 91:107–125. https://doi.org/10.1016/j.sajb.2014.01.002
Vettore AL, Da Silva FR, Kemper EL, Arruda P (2001) The libraries that made SUCEST. Genet Mol Biol 24:1–7. https://doi.org/10.1590/S1415-47572001000100002
Vicentini R, Bottcher A, Brito Mdos S, Dos Santos AB, Creste S, Landell MG et al (2015) Large-scale transcriptome analysis of two sugarcane genotypes contrasting for lignin content. PLoS One 10(8):e0134909. https://doi.org/10.1371/journal.pone.0134909
Vignols F, Rigau J, Torres MA, Puigdomenech P (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7:407–416
Voelker SL, Lachenbruch B, Meinzer FC, Jourdes M, Ki C, Patten AM et al (2010) Antisense down-regulation of 4CL expression alters lignification, tree growth, and sac-charification potential of field-grown poplar. Plant Physiol 154:874–886. https://doi.org/10.1104/pp.110.159269
Walker AM, Hayes RP, Youn B, Vermerris W, Sattler SE, Kang C (2013) Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme a-dependent transferases and synthases. Plant Physiol 162(2):640–651. https://doi.org/10.1104/pp.113.217836
Wang GF, Balint-Kurti PJ (2016) Maize Homologs of CCoAOMT and HCT, two key enzymes in lignin biosynthesis, form complexes with the NLR Rp1 protein to modulate the defense response. Plant Physiol 171(3):2166–2177. https://doi.org/10.1104/pp.16.00224
Wang JP, Naik PP, Chen H-C, Shi R, Lin CY, Liu J et al (2014) Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26:894–914. https://doi.org/10.1105/tpc.113.120881
Wang H, Li K, Hu X, Liu Z, Wu Y, Huang C (2016) Genome-wide association analysis of forage quality in maize mature stalk. BMC Plant Biol 16:227. https://doi.org/10.1186/s12870-016-0919-9
Watts KT, Mijts BN, Lee PC, Manning AJ, Schmidt-Dannert C (2006) Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem Biol 13:1317–1326. https://doi.org/10.1016/j.chembiol.2006.10.008
Weng JK, Akiyama T, Bonawitz ND, Li X, Ralph J, Chapple C (2010) Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophytes Selaginella and flowering plants. Plant Cell 22:1033–1045. https://doi.org/10.1105/tpc.109.073528
Widiez T, Hartman TG, Dudai N, Yan Q, Lawton M, Havkin-Frenkel D, Belanger FC (2011) Functional characterization of two new members of the caffeoyl CoA O-methyltransferase-like gene family from Vanilla planifolia reveals a new class of plastid-localized O-methyltransferases. Plant Mol Biol 76:475–488. https://doi.org/10.1007/s11103-011-9772-2
Xiong W, Wu Z, Liu Y, Li Y, Su K, Bai Z et al (2019) Mutation of 4-coumarate: coenzyme A ligase 1 gene afects lignin biosynthesis and increases the cell wall digestibility in maize brown midrib5 mutants. Biotechnol Biofuels 12:82. https://doi.org/10.1186/s13068-019-1421-z.158
Xu Z, Zhang D, Hu J, Zhou X, Ye X, Reichel KL et al (2009) Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinform 10(Suppl 1):S3. https://doi.org/10.1186/1471-2105-10-S11-S3
Xu B, Escamilla-Treviño LL, Sathitsuksanoh N, Shen Z, Shen H, Zhang YH et al (2011) Silencing of 4-coumarate: coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol 192:611–625. https://doi.org/10.1111/j.1469-8137.2011.03830.x
Yang Q, He Y, Kabahuma M, Chaya T, Kelly A, Borrego E et al (2017) A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat Genet 49(9):1364–1372. https://doi.org/10.1038/ng.3919
Ye ZH, Kneusel RE, Matern U, Varner JE (1994) An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell 6(10):1427–1439
Youn B, Camacho R, Moinuddin SGA, Lee C, Davin LB, Lewis NG, Kang C (2006) Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4. Org Biomol Chem 4:1687–1697. https://doi.org/10.1039/b601672c
Yu CS, Cheng CW, Su WC, Chang KC, Huang SW, Hwang JK, Lu CH (2014) CELLO2GO: a web server for protein subcellular localization prediction with functional gene ontology annotation. PLoS One 9(6):e99368. https://doi.org/10.1371/journal.pone.0099368
Yuan W, Jiang T, Du K, Chen H, Cao Y, Xie J, Li M, Carr JP, Wu B, Fan Z, Zhou T (2019) Maize phenylalanine ammonia-lyases contribute to resistance to Sugarcane mosaic virus infection, most likely through positive regulation of salicylic acid accumulation. Mol Plant Pathol 10:1365–1378. https://doi.org/10.1111/mpp.12817
Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L et al (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140(3):972–983. https://doi.org/10.1104/pp.105.073007
Zhang G, Zhang Y, Xu J, Niu X, Qi J, Tao A et al (2014) The CCoAOMT1 gene from jute (Corchorus capsularis L.) is involved in lignin biosynthesis in Arabidopsis thaliana. Gene 546:398–402. https://doi.org/10.1016/j.gene.2014.05.011
Zhang J, Zhang X, Tang H, Zhang Q, Hua X, Ma X et al (2018) Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat Genet 50:1565–1573. https://doi.org/10.1038/s41588-018-0237-2
Zhong R, Morrison WH, Himmelsbach DS, Poole FL, Ye Z-H (2000) Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol 124:563–577
Zhou R, Jackson L, Shadle G, Nakashima J, Temple S, Chen F, Dixon RA (2010) Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula. Proc Natl Acad Sci 107:17803–17808. https://doi.org/10.1073/pnas.1012900107
Acknowledgements
The authors would like to thank to Luiza da Silva for technical support and to Ana Coelho and Ana Giannini for critical review of the manuscript. This work is part of the Ph.D. thesis of José Nicomedes Júnior and Thais Felix-Cordeiro. DJM was supported by post-doctoral fellowship CAPES-PNPD and Petrobras. TFS, RL, and FPC were supported by post-doctoral fellowship CAPES-PNPD. JPF, CRF, and RL were supported by post-doctoral fellowship Petrobras. TFC was supported by CAPES PhD fellowship. LB, IB, TCS, JF, and KE were recipient of CNPq PIBIC fellowship.
Funding
This study was funded by Grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq (Grant number 474911/2012-8), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—CAPES (PNPD N°02691/09-4 - MEC / CAPES), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro—FAPERJ (E26/111.234/2014), and Petrobras (Grant number ANP 01070-0050.0076123.12.9) to GSM.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Authors declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jardim-Messeder, D., da Franca Silva, T., Fonseca, J.P. et al. Identification of genes from the general phenylpropanoid and monolignol-specific metabolism in two sugarcane lignin-contrasting genotypes. Mol Genet Genomics 295, 717–739 (2020). https://doi.org/10.1007/s00438-020-01653-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01653-1