Abstract
Purpose
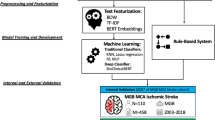
The highly structured nature of medical reports makes them feasible for automated large-scale patient identification. This study aimed to develop a natural language processing (NLP) model to retrospectively retrieve patients with presence and history of carotid stenosis (CS) using their ultrasound reports.
Methods
Ultrasound reports from our institution between January 2016 and December 2017 were selected. To process the texts, we developed a parser to divide the raw text into fields. For baseline method, we used bag-of-n-grams and term frequency inverse document frequency as the features and used linear classifiers. Logistic regression was performed as the baseline model. Convolution and recurrent neural networks (CNN; RNN) with attention mechanism were applied to the dataset to improve the classification accuracy.
Results
We had 1220 ultrasound reports for training and 307 for testing, totaling to 1527 reports. For predicting history of CS, both CNN and RNN-attention models had a significantly higher specificity than logistic regression. In addition, RNN-attention also had a significantly higher F1 score and accuracy. For predicting presence of carotid stenosis, all models achieved above 93% accuracy. RNN-attention achieved a 95.4% accuracy, although the difference with logistic regression was not statistically significant. RNN-attention had a statistically significant higher specificity than logistic regression.
Conclusions
We developed linear, CNN, and RNN models to predict history and presence of CS from ultrasound reports. We have demonstrated NLP to be an efficient, accurate approach for large-scale retrospective patient identification, with applications in long-term follow-up of patients and clinical research studies.
Key Points
• Natural language processing models using both linear classifiers and neural networks can achieve a good performance, with an overall accuracy above 90% in predicting history and presence of carotid stenosis.
• Convolution and recurrent neural networks, especially with additional features including field awareness and attention mechanism, have superior performance than traditional linear classifiers.
• NLP is shown to be an efficient approach for large-scale retrospective patient identification, with applications in long-term follow-up of patients and further clinical research studies.





Similar content being viewed by others
Abbreviations
- CI:
-
Confidence interval
- CNN:
-
Convolutional neural network
- CS:
-
Carotid stenosis
- GATE:
-
General Architecture for Text Engineering
- ICD-9:
-
International Classification of Diseases, Ninth Revision
- LSTM:
-
Long-short-term memory cells
- NASCET:
-
The North American Symptomatic Carotid Endarterectomy Trial
- NLP:
-
Natural language processing
- RNN:
-
Recurrent neural network
- ROC:
-
Receiver operating characteristics
- US:
-
Ultrasound
References
Brott TG, Halperin JL, Abbara S et al (2013) 2011 ASA/ACCF/AHA/AANN/AANS/ACR/ASNR/CNS/SAIP/SCAI/SIR/SNIS/SVM/SVS guideline on the management of patients with extracranial carotid and vertebral artery disease: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, and the American Stroke Association, American Association of Neuroscience Nurses, American Association of Neurological Surgeons, American College of Radiology, American Society of Neuroradiology, Congress of Neurological Surgeons, Society of Atherosclerosis Imaging and Prevention, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of NeuroInterventional Surgery, Society for Vascular Medicine, and Society for Vascular Surgery. Developed in collaboration with the American Academy of Neurology and Society of Cardiovascular Computed Tomography. Catheter Cardiovasc Interv 81:E76–E123
(1991) Clinical alert: benefit of carotid endarterectomy for patients with high-grade stenosis of the internal carotid artery. National Institute of Neurological Disorders and Stroke Stroke and Trauma Division. North American Symptomatic Carotid Endarterectomy Trial (NASCET) investigators. Stroke 22:816–817
Abbott AL, Bladin CF, Levi CR, Chambers BR (2007) What should we do with asymptomatic carotid stenosis? Int J Stroke 2:27–39
Grant EG, Benson CB, Moneta GL et al (2003) Carotid artery stenosis: gray-scale and Doppler US diagnosis--Society of Radiologists in Ultrasound Consensus Conference. Radiology 229:340–346
Bazarian JJ, Veazie P, Mookerjee S, Lerner EB (2006) Accuracy of mild traumatic brain injury case ascertainment using ICD-9 codes. Acad Emerg Med 13:31–38
Benesch C, Witter DM Jr, Wilder AL, Duncan PW, Samsa GP, Matchar DB (1997) Inaccuracy of the International Classification of Diseases (ICD-9-CM) in identifying the diagnosis of ischemic cerebrovascular disease. Neurology 49:660–664
Jensen PB, Jensen LJ, Brunak S (2012) Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 13:395–405
Pons E, Braun LM, Hunink MG, Kors JA (2016) Natural language processing in radiology: a systematic review. Radiology 279:329–343
Lakhani P, Kim W, Langlotz CP (2012) Automated detection of critical results in radiology reports. J Digit Imaging 25:30–36
Rink B, Roberts K, Harabagiu S et al (2013) Extracting actionable findings of appendicitis from radiology reports using natural language processing. AMIA Jt Summits Transl Sci Proc 2013:221–221
Yetisgen-Yildiz M, Gunn ML, Xia F, Payne TH (2013) A text processing pipeline to extract recommendations from radiology reports. J Biomed Inform 46:354–362
Chen Y (2015) Convolutional neural network for sentence classification. UWSpace, University of Waterloo, Ontario, Canada
Kim Y (2014) Convolutional neural networks for sentence classification. arXiv:14085882
Joulin A, Grave E, Bojanowski P, Mikolov T (2016) Bag of tricks for efficient text classification. arXiv:160701759
Tang D, Qin B, Liu T (2015) Document modeling with gated recurrent neural network for sentiment classification. Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, Lisbon, pp 1422–1432
Bahdanau D, Cho K, Bengio Y (2014) Neural machine translation by jointly learning to align and translate. arXiv:14090473
Danforth KN, Early MI, Ngan S, Kosco AE, Zheng C, Gould MK (2012) Automated identification of patients with pulmonary nodules in an integrated health system using administrative health plan data, radiology reports, and natural language processing. J Thorac Oncol 7:1257–1262
Blei DM, Ng AY, Jordan MI (2002) Latent Dirichlet allocation. Advances in neural information processing systems, pp 601-608
Blei DM, Ng AY, Jordan MI (2003) Latent dirichlet allocation. J Mach Learn Res 3:993–1022
Hoffman M, Bach FR, Blei DM (2010) Online learning for latent Dirichlet allocation. Advances in neural information processing systems, pp 856-864
Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ (2000) Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc 7:593–604
Dublin S, Baldwin E, Walker RL et al (2013) Natural language processing to identify pneumonia from radiology reports. Pharmacoepidemiol Drug Saf 22:834–841
Elkin PL, Froehling D, Wahner-Roedler D et al (2008) NLP-based identification of pneumonia cases from free-text radiological reports. AMIA Ann Symp Proc 2008:172–176
Chapman BE, Lee S, Kang HP, Chapman WW (2011) Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm. J Biomed Inform 44:728–737
Yu S, Kumamaru KK, George E et al (2014) Classification of CT pulmonary angiography reports by presence, chronicity, and location of pulmonary embolism with natural language processing. J Biomed Inform 52:386–393
Cheng LT, Zheng J, Savova GK, Erickson BJ (2010) Discerning tumor status from unstructured MRI reports--completeness of information in existing reports and utility of automated natural language processing. J Digit Imaging 23:119–132
Jain NL, Friedman C (1997) Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp 829–833
Maehara CK, Silverman SG, Lacson R, Khorasani R (2014) Renal masses detected at abdominal CT: radiologists’ adherence to guidelines regarding management recommendations and communication of critical results. AJR Am J Roentgenol 203:828–834
Pham AD, Neveol A, Lavergne T et al (2014) Natural language processing of radiology reports for the detection of thromboembolic diseases and clinically relevant incidental findings. BMC Bioinformatics 15:266
Savova GK, Masanz JJ, Ogren PV et al (2010) Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 17:507–513
Swartz J, Koziatek C, Theobald J, Smith S, Iturrate E (2017) Creation of a simple natural language processing tool to support an imaging utilization quality dashboard. Int J Med Inform 101:93–99
Zhang Y, Ding DY, Qian T, Manning CD, Langlotz CP (2018) Learning to summarize radiology findings. arXiv:1809.04698
Dutta S, Long WJ, Brown DF, Reisner AT (2013) Automated detection using natural language processing of radiologists recommendations for additional imaging of incidental findings. Ann Emerg Med 62:162–169
Sistrom CL, Dreyer KJ, Dang PP et al (2009) Recommendations for additional imaging in radiology reports: multifactorial analysis of 5.9 million examinations. Radiology 253:453–461
O'Connor SD, Silverman SG, Ip IK, Maehara CK, Khorasani R (2013) Simple cyst-appearing renal masses at unenhanced CT: can they be presumed to be benign? Radiology 269:793–800
Percha B, Nassif H, Lipson J, Burnside E, Rubin D (2012) Automatic classification of mammography reports by BI-RADS breast tissue composition class. J Am Med Inform Assoc 19:913–916
Chen MC, Ball RL, Yang L et al (2018) Deep learning to classify radiology free-text reports. Radiology 286:845–852
Brott TG, Halperin JL, Abbara S et al (2011) 2011 ASA/ACCF/AHA/AANN/AANS/ACR/ASNR/CNS/SAIP/SCAI/SIR/SNIS/SVM/SVS guideline on the management of patients with extracranial carotid and vertebral artery disease. Stroke 42:e464–e540
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Ajay Malhotra.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Experimental
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 16 kb)
Rights and permissions
About this article
Cite this article
Wu, X., Zhao, Y., Radev, D. et al. Identification of patients with carotid stenosis using natural language processing. Eur Radiol 30, 4125–4133 (2020). https://doi.org/10.1007/s00330-020-06721-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-06721-z