Abstract
Objective
To reveal the systematic response of neutrophils to sepsis and to study the hub lncRNAs in sepsis.
Materials and methods
Neutrophils taken from the femur and tibia of male C57 BL/6 mice were used in this study. And neutrophils were treated for 0 h, 0.5 h, 1 h, and 4 h with or without 1 µg/mL lipopolysaccharide (LPS) for further chip detection. In addition, cecal ligation and perforation were used to simulate sepsis. Here, we used different bioinformatics analyses, including differential expression analysis, weighted gene co-expression network analysis (WGCNA), and gene regulatory network analysis, to analyze the systemic response of neutrophils to sepsis.
Results
We identified nine modules and found hub lncRNAs in each module. The blue and pink modules were closely related to the inflammatory state of sepsis. Some hub lncRNAs (NONMMUT005259, KnowTID_00004196, and NR_003507) may have functions related to the inflammatory state in sepsis.
Conclusions
Based on a new biological approach, our research results revealed the systemic-level response of neutrophils to sepsis and identified several hub lncRNAs with potential regulatory effects on this condition.






Similar content being viewed by others
References
Lelubre C, Vincent JL. Mechanisms and treatment of organ failure in sepsis. Nat Rev Nephrol. 2018;14(7):417–27.
Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA. 2016;315(8):801–10.
Venet F, Monneret G. Advances in the understanding and treatment of sepsis-induced immunosuppression. Nat Rev Nephrol. 2018;14(2):121–37.
Wiersinga WJ, Leopold SJ, Cranendonk DR, van der Poll T. Host innate immune responses to sepsis. Virulence. 2014;5(1):36–44.
Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. 2013;13(12):862–74.
Leliefeld PH, Wessels CM, Leenen LP, Koenderman L, Pillay J. The role of neutrophils in immune dysfunction during severe inflammation. Crit Care. 2016;20:73.
Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. 2017;37(1):3–9.
Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–325.
Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62.
Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–69.
Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription. Sci Adv. 2017;3(9):eaao2110.
Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genom Proteom Bioinform. 2017;15(3):177–86.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41.
Liu N, Pan T. RNA epigenetics. Transl Res. 2015;165(1):28–35.
Mo XB, Wu LF, Zhu XW, Xia W, Wang L, He P, et al. Identification and evaluation of lncRNA and mRNA integrative modules in human peripheral blood mononuclear cells. Epigenomics. 2017;9(7):943–54.
Liu W, Li L, Ye H, Tu W. Weighted gene co-expression network analysis in biomedicine research. Sheng Wu Gong Cheng Xue Bao. 2017;33(11):1791–801.
Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–504.
Ahn R, Gupta R, Lai K, Chopra N, Arron ST, Liao W. Network analysis of psoriasis reveals biological pathways and roles for coding and long non-coding RNAs. BMC Genom. 2016;17(1):841.
Miao X, Luo Q, Zhao H, Qin X. Co-expression analysis and identification of fecundity-related long non-coding RNAs in sheep ovaries. Sci Rep. 2016;6:39398.
Zhao G, Fu Y, Su Z, Wu R. How long non-coding RNAs and MicroRNAs mediate the endogenous RNA network of head and neck squamous cell carcinoma: a comprehensive analysis. Cell Physiol Biochem. 2018;50(1):332–41.
Giulietti M, Occhipinti G, Principato G, Piva F. Weighted gene co-expression network analysis reveals key genes involved in pancreatic ductal adenocarcinoma development. Cell Oncol (Dordr). 2016;39(4):379–88.
Zhou XG, Huang XL, Liang SY, Tang SM, Wu SK, Huang TT, et al. Identifying miRNA and gene modules of colon cancer associated with pathological stage by weighted gene co-expression network analysis. Onco Targets Ther. 2018;11:2815–30.
Pei G, Chen L, Zhang W. WGCNA application to proteomic and metabolomic data analysis. Methods Enzymol. 2017;585:135–58.
Godini R, Fallahi H, Ebrahimie E. Network analysis of inflammatory responses to sepsis by neutrophils and peripheral blood mononuclear cells. PLoS ONE. 2018;13(8):e0201674.
Chen L, Zhang YH, Wang S, Zhang Y, Huang T, Cai YD. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE. 2017;12(9):e0184129.
Zhang C, Peng L, Zhang Y, Liu Z, Li W, Chen S, et al. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med Oncol. 2017;34(6):101.
Liu X, Hu AX, Zhao JL, Chen FL. Identification of key gene modules in human osteosarcoma by co-expression analysis weighted gene co-expression network analysis (WGCNA). J Cell Biochem. 2017;118(11):3953–9.
Fatima A, Connaughton RM, Weiser A, Murphy AM, O’Grada C, Ryan M, et al. Weighted gene co-expression network analy-sis identifiesgender specific modules and hub genes related to metabolism and inflammation in response to an acute lipid chal-lenge. Mol Nutr Food Res. 2018;62(2):1–23.
Rogobete AF, Sandesc D, Papurica M, Stoicescu ER, Popovici SE, Bratu LM, et al. The influence of metabolic imbalances and oxidative stress on the outcome of critically ill polytrauma patients: a review. Burn Trauma. 2017;5:8.
Wang X, Qin W, Sun B. New strategy for sepsis: targeting a key role of platelet-neutrophil interaction. Burn Trauma. 2014;2(3):114–20.
Castelo-Branco G, Bonetti A. Birth, coming of age and death: the intriguing life of long noncoding RNAs. Semin Cell Dev Biol. 2018;79:143–52.
Othumpangat S, Bryan NB, Beezhold DH, Noti JD. Upregulation of miRNA-4776 in influenza virus infected bronchial epithelial cells is associated with downregulation of NFKBIB and increased viral survival. Viruses. 2017;9(5):1–18.
Sun SC. The noncanonical NF-κB pathway. Immunol Rev. 2012;246(1):125–40.
Häcker H, Karin M. Is NF-kappaB2/p100 a direct activator of programmed cell death. Cancer Cell. 2002;2(6):431–3.
Majoros A, Platanitis E, Kernbauer-Hölzl E, Rosebrock F, Müller M, Decker T. Canonical and non-canonical aspects of JAK-STAT signaling: lessons from interferons for cytokine responses. Front Immunol. 2017;8:29.
Paul A, Tang TH, Ng SK. Interferon regulatory factor 9 structure and regulation. Front Immunol. 2018;9:1831.
Blaszczyk K, Nowicka H, Kostyrko K, Antonczyk A, Wesoly J, Bluyssen HA. The unique role of STAT2 in constitutive and IFN-induced transcription and antiviral responses. Cytokine Growth Factor Rev. 2016;29:71–81.
Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12(6):399–414.
Zhang L, Pagano JS. Structure and function of IRF-7. J Interferon Cytokine Res. 2002;22(1):95–101.
Ersing I, Bernhardt K, Gewurz BE. NF-κB and IRF7 pathway activation by Epstein-Barr virus latent membrane protein 1. Viruses. 2013;5(6):1587–606.
Acknowledgements
This study was supported by the National Natural Science Foundation of China, Nos. 81471903, and 81772135; by the Jiangsu Natural Science Foundation, No. BE2017695.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Responsible Editor: John Di Battista.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11_2020_1323_MOESM1_ESM.tif
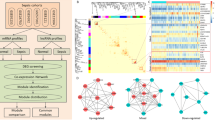
Supplementary file1 (TIF 983 kb) Weighted co-expression gene network analysis. (a) Sample cluster analysis. (b) Number of mRNAs and lncRNAs in each module. (c) Eigengene adjacency heatmap. Colors represent correlation between two modules: blue, low correlation; red, high correlation
11_2020_1323_MOESM2_ESM.tif
Supplementary file2 (TIF 1179 kb) GO enrichment of DEGs in the turquoise (a), green (b), and brown (c) modules. KEGG annotation of gene clusters in the turquoise (d), green (e), and brown (f) modules. Colors represent term enrichment: purple, low enrichment; red, high enrichment
11_2020_1323_MOESM3_ESM.tif
Supplementary file3 (TIF 2609 kb) Module lncRNA-mRNA-Net of hub genes in the turquoise (a), green (b), and brown (c) modules. Module lncRNA-mRNA-Pathway-Net of hub genes in the turquoise (d), green (e), and brown (f) modules. Green circles represent mRNAs, red triangles represent lncRNAs, and gray polygons represent pathways. The size of these graphs represents the level of intramodular connectivity of hub genes in the network
Rights and permissions
About this article
Cite this article
Huang, J., Sun, R. & Sun, B. Identification and evaluation of hub mRNAs and long non-coding RNAs in neutrophils during sepsis. Inflamm. Res. 69, 321–330 (2020). https://doi.org/10.1007/s00011-020-01323-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-020-01323-3