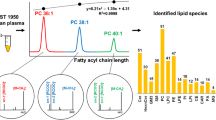
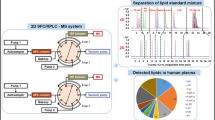
Abstract
Ultrahigh-performance supercritical fluid chromatography–mass spectrometry (UHPSFC/MS) has a great potential for the high-throughput lipidomic quantitation of biological samples; therefore, the full optimization and method validation of UHPSFC/MS is compared here with ultrahigh-performance liquid chromatography–mass spectrometry (UHPLC/MS) in hydrophilic interaction liquid chromatography (HILIC) mode as the second powerful technique for the lipid class separation. First, the performance of six common extraction protocols is investigated, where the Folch procedure yields the best results with regard to recovery rate, matrix effect, and precision. Then, the full optimization and analytical validation for eight lipid classes using UHPSFC/MS and HILIC-UHPLC/MS methods are performed for the same sample set and applied for the lipidomic characterization of pooled samples of human plasma, human serum, and NIST SRM 1950 human plasma. The choice of appropriate internal standards (IS) for individual lipid classes has a key importance for reliable quantitative workflows illustrated by the selectivity while validation and the calculation of the quantitation error using multiple internal standards per lipid class. Validation results confirm the applicability of both methods, but UHPSFC/MS provides some distinct advantages, such as the successful separation of both non-polar and polar lipid classes unlike to HILIC-UHPLC/MS, shorter total run times (8 vs. 10.5 min), and slightly higher robustness. Various types of correlations between methods (UHPSFC/MS and HILIC-UHPLC/MS), biological material (plasma and serum), IS (laboratory and commercially mixtures), and literature data on the standard reference material show the intra- and inter-laboratory comparison in the quantitation of lipid species from eight lipid classes, the concentration differences in serum and plasma as well as the applicability of non-commercially available internal standard mixtures for lipid quantitation.





Similar content being viewed by others
References
Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610.
Han X. Lipidomics for studying metabolism. Nat Rev Endocrinol. 2016;12:668.
Quehenberger O, Dennis EA. The human plasma lipidome. N Engl J Med. 2011;365:1812–23.
Stephenson DJ, Hoeferlin LA, Chalfant CE. Lipidomics in translational research and the clinical significance of lipid-based biomarkers. Transl Res. 2017;189:13–29.
Wolrab D, Jirásko R, Chocholoušková M, Peterka O, Holčapek M. Oncolipidomics: mass spectrometric quantitation of lipids in cancer research. TrAC Trends Anal Chem. 2019;120:115480.
Chua EC-P, Shui G, Lee IT-G, Lau P, Tan L-C, Yeo S-C, et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc Natl Acad Sci U S A. 2013;110:14468.
Begum H, Li B, Shui G, Cazenave-Gassiot A, Soong R, Ong RT-H, et al. Discovering and validating between-subject variations in plasma lipids in healthy subjects. Sci Rep. 2016;6:19139.
Sales S, Graessler J, Ciucci S, Al-Atrib R, Vihervaara T, Schuhmann K, Kauhanen D, Sysi-Aho M, Bornstein SR, Bickle M, Cannistraci CV, Ekroos K, Shevchenko A. Gender, contraceptives and individual metabolic predisposition shape a healthy plasma lipidome Sci Rep 2016;6:27710.
Bowden JA, Heckert A, Ulmer CZ, Jones CM, Koelmel JP, Abdullah L, et al. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950–Metabolites in Frozen Human Plasma. J Lipid Res. 2017;58:2275–88.
Burla B, Arita M, Arita M, Bendt AK, Cazenave-Gassiot A, Dennis EA, et al. MS-based lipidomics of human blood plasma: a community-initiated position paper to develop accepted guidelines. J Lipid Res. 2018;59:2001–17.
Holčapek M, Liebisch G, Ekroos K. Lipidomic analysis. Anal Chem. 2018;90:4249–57.
Rampler E, Schoeny H, Mitic BM, El Abiead Y, Schwaiger M, Koellensperger G. Simultaneous non-polar and polar lipid analysis by on-line combination of HILIC, RP and high resolution MS. Analyst. 2018;143:1250–8.
Cífková E, Holčapek M, Lísa M, Vrána D, Melichar B, Študent V. Lipidomic differentiation between human kidney tumors and surrounding normal tissues using HILIC-HPLC/ESI–MS and multivariate data analysis. J Chromatogr B. 2015;1000:14–21.
Triebl A, Trötzmüller M, Hartler J, Stojakovic T, Köfeler HC. Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples. J Chromatogr B. 2017;1053:72–80.
Lísa M, Holčapek M. High-throughput and comprehensive lipidomic analysis using ultrahigh-performance supercritical fluid chromatography–mass spectrometry. Anal Chem. 2015;87:7187–95.
Lísa M, Cífková E, Khalikova M, Ovčačíková M, Holčapek M. Lipidomic analysis of biological samples: comparison of liquid chromatography, supercritical fluid chromatography and direct infusion mass spectrometry methods. J Chromatogr A. 2017;1525:96–108.
Kočová Vlčková H, Pilařová V, Svobodová P, Plíšek J, Švec F, Nováková L. Current state of bioanalytical chromatography in clinical analysis. Analyst. 2018;143:1305–25.
Bamba T, Shimonishi N, Matsubara A, Hirata K, Nakazawa Y, Kobayashi A, et al. High throughput and exhaustive analysis of diverse lipids by using supercritical fluid chromatography-mass spectrometry for metabolomics. J Biosci Bioeng. 2008;105:460–9.
Desfontaine V, Losacco GL, Gagnebin Y, Pezzatti J, Farrell WP, González-Ruiz V, et al. Applicability of supercritical fluid chromatography – mass spectrometry to metabolomics. I – optimization of separation conditions for the simultaneous analysis of hydrophilic and lipophilic substances. J Chromatogr A. 2018;1562:96–107.
Dispas A, Jambo H, André S, Tyteca E, Hubert P. Supercritical fluid chromatography: a promising alternative to current bioanalytical techniques. Bioanalysis. 2018;10:107–24.
West C. Current trends in supercritical fluid chromatography. Anal Bioanal Chem. 2018;410:6441–57.
Lesellier E, West C. The many faces of packed column supercritical fluid chromatography – a critical review. J Chromatogr A. 2015;1382:2–46.
Liebisch G, Ahrends R, Arita M, Arita M, Bowden JA, Ejsing CS, et al. Lipidomics standards initiative C. Lipidomics needs more standardization. Nature Metabolism. 2019;1:745–7.
McCalley DV. A study of column equilibration time in hydrophilic interaction chromatography. J Chromatogr A. 2018;1554:61–70.
Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509.
Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–46.
Gil A, Zhang W, Wolters JC, Permentier H, Boer T, Horvatovich P, et al. One- vs two-phase extraction: re-evaluation of sample preparation procedures for untargeted lipidomics in plasma samples. Anal Bioanal Chem. 2018;410:5859–70.
Löfgren L, Forsberg G-B, Ståhlman M. The BUME method: a new rapid and simple chloroform-free method for total lipid extraction of animal tissue. Sci Rep. 2016;6:27688.
Shibusawa Y, Yamakawa Y, Noji R, Yanagida A, Shindo H, Ito Y. Three-phase solvent systems for comprehensive separation of a wide variety of compounds by high-speed counter-current chromatography. J Chromatogr A. 2006;1133:119–25.
U.S. Department of Health and Human Services Food and Drug Administration, Draft Guidance for Industry: Bioanalytical Method Validation. 2018. https://www.fda.gov/files/drugs/published/Bioanalytical-Method-Validation-Guidance-for-Industry.pdf. Accessed 15 Oct 2019.
European Medicines Agency, EMEA/CHMP/EWP/192217/2009 Rev. 1 Corr. 2, Guideline on bioanalytical method validation. 2011. https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-bioanalytical-method-validation_en.pdf. Accessed 15 Oct 2019.
European Medicines Agency,EMA/CHMP/ICH/172948/2019, ICH guideline M10 on bioanalytical method validation Step 2b. 2019. https://www.ema.europa.eu/en/documents/scientific-guideline/draft-ich-guideline-m10-bioanalytical-method-validation-step-2b_en.pdf. Accessed 15 Oct 2019.
Kadian N, Raju KSR, Rashid M, Malik MY, Taneja I, Wahajuddin M. Comparative assessment of bioanalytical method validation guidelines for pharmaceutical industry. J Pharm Biomed Anal. 2016;126:83–97.
González O, Blanco ME, Iriarte G, Bartolomé L, Maguregui MI, Alonso RM. Bioanalytical chromatographic method validation according to current regulations, with a special focus on the non-well defined parameters limit of quantification, robustness and matrix effect. J Chromatogr A. 2014;1353:10–27.
Rawski RI, Sanecki PT, Kijowska KM, Skital PM, Saletnik DE. Regression analysis in analytical chemistry. Determination and validation of linear and quadratic regression dependencies : research article. South Afr J Chem. 2016;69:166–73.
Andrade JM, Gómez-Carracedo MP. Notes on the use of Mandel's test to check for nonlinearity in laboratory calibrations. Anal Methods. 2013;5:1145–9.
Cajka T, Smilowitz JT, Fiehn O. Validating quantitative untargeted lipidomics across nine liquid chromatography–high-resolution mass spectrometry platforms. Anal Chem. 2017;89:12360–8.
Chocholoušková M, Jirásko R, Vrána D, Gatěk J, Melichar B, Holčapek M. Reversed phase UHPLC/ESI-MS determination of oxylipins in human plasma: a case study of female breast cancer. Anal Bioanal Chem. 2019;411:1239–51.
Lee DY, Kind T, Yoon Y-R, Fiehn O, Liu K-H. Comparative evaluation of extraction methods for simultaneous mass-spectrometric analysis of complex lipids and primary metabolites from human blood plasma. Anal Bioanal Chem. 2014;406:7275–86.
Triebl A, Burla B, Selvalatchmanan J, Oh J, Tan SH, Chan MY, Mellett NA, Meikle PJ, Torta F, Wenk MR. Shared reference materials harmonize lipidomics across MS-based detection platforms and laboratories. J Lipid Res. 2020;61:105–15.
Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51:3299–305.
Acknowledgments
We would like to acknowledge the help of the group of Bohuslav Melichar (University Hospital Olomouc, Czech Republic) with the sample collection and other people involved in the sample processing from University of Pardubice (Vladimíra Nováková Mužáková, Jana Kňavová, and Iva Fousová).
Funding
This work was supported by the grant project no. 18-12204S funded by the Czech Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The study was approved by the ethical committee at University Hospital Olomouc, Czech Republic, and all healthy volunteers signed informed consent.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Published in the topical collection Current Progress in Lipidomics with guest editors Michal Holčapek, Gerhard Liebisch, and Kim Ekroos.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wolrab, D., Chocholoušková, M., Jirásko, R. et al. Validation of lipidomic analysis of human plasma and serum by supercritical fluid chromatography–mass spectrometry and hydrophilic interaction liquid chromatography–mass spectrometry. Anal Bioanal Chem 412, 2375–2388 (2020). https://doi.org/10.1007/s00216-020-02473-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02473-3