Abstract
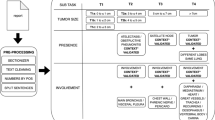
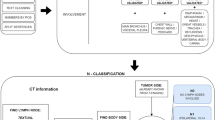
Reports are the standard way of communication between the radiologist and the referring clinician. Efforts are made to improve this communication by, for instance, introducing standardization and structured reporting. Natural Language Processing (NLP) is another promising tool which can improve and enhance the radiological report by processing free text. NLP as such adds structure to the report and exposes the information, which in turn can be used for further analysis. This paper describes pre-processing and processing steps and highlights important challenges to overcome in order to successfully implement a free text mining algorithm using NLP tools and machine learning in a small language area, like Dutch. A rule-based algorithm was constructed to classify T-stage of pulmonary oncology from the original free text radiological report, based on the items tumor size, presence and involvement according to the 8th TNM classification system. PyContextNLP, spaCy and regular expressions were used as tools to extract the correct information and process the free text. Overall accuracy of the algorithm for evaluating T-stage was 0,83 in the training set and 0,87 in the validation set, which shows that the approach in this pilot study is promising. Future research with larger datasets and external validation is needed to be able to introduce more machine learning approaches and perhaps to reduce required input efforts of domain-specific knowledge. However, a hybrid NLP approach will probably achieve the best results.


Similar content being viewed by others
References
McGinty GB, Allen B, Geis JR, Wald C: IT infrastructure in the era of imaging 3.0. J Am Coll Radiol 11:1197–1204, 2014
Brierley J, Gospodarowicz MK, Wittekind C Eds: TNM classification of malignant tumours, 8th edition. Chichester: John Wiley & Sons Inc., 2017
Puts S, Nobel JM: Medical narrative to structure: maastroclinic/medstruct. maastroclinic, 2019
Krupinski EA, Hall ET, Jaw S, Reiner B, Siegel E: Influence of radiology report format on reading time and comprehension. J Digit Imaging 25:63–69, 2012
Pons E, Braun LMM, Hunink MGM, Kors JA: Natural language processing in radiology: A systematic review. Radiology 279:329–343, 2016
Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG: Mayo clinical text analysis and knowledge extraction system (cTAKES): Architecture, component evaluation and applications. J Am Med Inform Assoc 17:507–513, 2010
Cornet R, van Eldik A, de Keizer N: Inventory of tools for Dutch clinical language processing. Stud Health Technol Inform 180:245–249, 2012
Nguyen AN, Lawley MJ, Hansen DP, Bowman RV, Clarke BE, Duhig EE, Colquist S: Symbolic rule-based classification of lung cancer stages from free-text pathology reports. J Am Med Inform Assoc 17:440–445, 2010
Castro SM, Tseytlin E, Medvedeva O, Mitchell K, Visweswaran S, Bekhuis T, Jacobson RT: Automated annotation and classification of BI-RADS assessment from radiology reports. J Biomed Inform 69:177–187, 2017
Pathak S, van Rossen J, Vijlbrief O, Geerdink J, Seifert C, van Keulen M: Automatic Structuring of Breast Cancer Radiology Reports for Quality Assurance. IEEE international conference on data mining workshops (ICDMW), Singapore, IEEE 2018(732–739):2018, 2018
Honnibal M, Montani I: Spacy 2: Natural language understanding with bloom embeddings, convolutional neural networks and incremental parsing. To appear: 7, 2017
Soldaini L, Goharian N: QuickUMLS: a fast, unsupervised approach for medical concept extraction. MedIR workshop, sigir, 2016. Available at http://ir.cs.georgetown.edu/downloads/quickumls.pdf. Accessed 6 May 2019.
Côté RA, Robboy S: Progress in medical information management. Systematized nomenclature of medicine (SNOMED). JAMA 243:756–762, 1980
Chapman BE, Lee S, Kang HP, Chapman WW: Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm. J Biomed Inform 44:728–737, 2011
Chapman WW, Hillert D, Velupillai S, Kvist M, Skeppstedt M, Chapman BE, Conway M, Tharp M, Mowery DL, Deleger L: Extending the NegEx lexicon for multiple languages. Stud Health Technol Inform 192:677–681, 2013
Afzal Z, Pons E, Kang N, Sturkenboom MC, Schuemie MJ, Kors JA: ContextD: An algorithm to identify contextual properties of medical terms in a Dutch clinical corpus. BMC Bioinformatics 15:373, 2014
Chapman WW: Extract context modifiers targeting clinical terms: Maastroclinic/pyConTextNLP 2019. Available at https://github.com/maastroclinic/pyConTextNLP. Accessed 19 June 2019.
Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJ, Groth P, Goble C, Grethe JS, Heringa J, ’t Hoen PA, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B: The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3:160018, 2016
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 25 kb)
Rights and permissions
About this article
Cite this article
Nobel, J.M., Puts, S., Bakers, F.C.H. et al. Natural Language Processing in Dutch Free Text Radiology Reports: Challenges in a Small Language Area Staging Pulmonary Oncology. J Digit Imaging 33, 1002–1008 (2020). https://doi.org/10.1007/s10278-020-00327-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-020-00327-z