Abstract
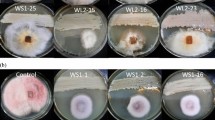
Aflatoxins are the most common mycotoxin contaminant reported in food and feed. Aflatoxin B1, the most toxic among different aflatoxins, is known to cause hepatocellular carcinoma in animals. Aspergillus flavus and A. parasiticus are the main producers of aflatoxins and are widely distributed in tropical countries. Even though several robust strategies have been in use to control aflatoxin contamination, the control at the pre-harvest level is primitive and incompetent. Therefore, the aim of the study was to isolate and identify the non-aflatoxigenic A. flavus and to delineate the molecular mechanism for the loss of aflatoxin production by the non-aflatoxigenic isolates. Eighteen non-aflatoxigenic strains were isolated from various biological sources using cultural and analytical methods. Among the 18 isolates, 8 isolates produced sclerotia and 17 isolates had type I deletion in norB-cypA region. The isolates were confirmed as A. flavus using gene-specific PCR and sequencing of the ITS region. Later, aflatoxin gene-specific PCR revealed that the defect in one or more genes has led to non-aflatoxigenic phenotype. The strain R9 had maximum defect, and genes avnA and verB had the highest frequency of defect among the non-aflatoxigenic strains. Further, qRT-PCR confirmed that the non-aflatoxigenic strains had high frequency of defect or downregulation in the late pathway genes compared to early pathway genes. Thus, these non-aflatoxigenic strains can be the potential candidates for an effective and proficient strategy for the control of pre-harvest aflatoxin contamination.




Similar content being viewed by others
References
Abbas HK, Shier WT, Horn BW, Weaver MA (2004a) Cultural methods for aflatoxin detection. J Toxicol Toxin Rev 23:295–315
Abbas HK, Zablotowicz RM, Weaver MA, Horn BW, Xie W, Shier WT (2004b) Comparison of cultural and analytical methods for determination of aflatoxin production by Mississippi Delta Aspergillus isolates. Can J Microbiol 50:193–199
Abbas HK, Weaver MA, Zablotowicz RM, Horn BW, Shier WT (2005) Relationships between aflatoxin production and sclerotia formation among isolates of Aspergillus section Flavi from the Mississippi Delta. Eur J Plant Pathol 112:283–287
Amaike S, Keller NP (2011) Aspergillus flavus. Annu Rev Phytopathol 49:107–133
Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS (2016) Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 44:W147–W153
Bayman P, Cotty PJ (1993) Genetic diversity in Aspergillus flavus: association with aflatoxin production and morphology. Can J Bot 71:23–31
Chang P-K, Horn BW, Dorner JW (2005) Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet Biol 42:914–923
Chang P-K, Ehrlich KC, Hua S-ST (2006) Cladal relatedness among Aspergillus oryzae isolates and Aspergillus flavus S and L morphotype isolates. Int J Food Microbiol 108:172–177
Chang P-K, Wilkinson JR, Horn BW, Yu J, Bhatnagar D, Cleveland TE (2007) Genes differentially expressed by Aspergillus flavus strains after loss of aflatoxin production by serial transfers. Appl Microbiol Biotechnol 77:917–925
Cotty PJ (1988) Aflatoxin and sclerotial production by Aspergillus flavus: influence of pH. Phytopathology 78:1250–1253
Cotty PJ (1989) Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology 79:808–814
Cotty PJ, Bayman P (1993) Competitive exclusion of a toxigenic strain of Aspergillus flavus by an atoxigenic strain. Phytopathology 83:1283–1287
Criseo G, Bagnara A, Bisignano G (2001) Differentiation of aflatoxin-producing and non-producing strains of Aspergillus flavus group. Lett Appl Microbiol 33:291–295
Criseo G, Racco C, Romeo O (2008) High genetic variability in non-aflatoxigenic A. flavus strains by using quadruplex PCR-based assay. Int J Food Microbiol 125:341–343
Davis ND, Iyer SK, Diener UL (1987) Improved method of screening for aflatoxin with a coconut agar medium. Appl Environ Microbiol 53:1593–1595
De Beeck MO, Lievens B, Busschaert P, Declerck S, Vangronsveld J, Colpaert JV (2014) Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS ONE 9:e97629
Degola F, Berni E, Dall’Asta C, Spotti E, Marchelli R, Ferrero I, Restivo FM (2007) A multiplex RT-PCR approach to detect aflatoxigenic strains of Aspergillus flavus. J Appl Microbiol 103:409–417
Donner M, Atehnkeng J, Sikora RA, Bandyopadhyay R, Cotty PJ (2010) Molecular characterization of atoxigenic strains for biological control of aflatoxins in Nigeria. Food Addit Contam 27:576–590
Dorner JW (2004) Biological control of aflatoxin contamination of crops. J Toxicol Toxin Rev 23:425–450
Ehrlich KC, Yu J, Cotty PJ (2005) Aflatoxin biosynthesis gene clusters and flanking regions. J Appl Microbiol 99:518–527
Färber P, Geisen R, Holzapfel WH (1997) Detection of aflatoxinogenic fungi in figs by a PCR reaction. Int J Food Microbiol 36:215–220
Hariprasad P, Vipin AV, Karuna S, Raksha RK, Venkateswaran G (2015) Natural aflatoxin uptake by sugarcane (Saccharum officinaurum L.) and its persistence in jaggery. Environ Sci Pollut Res 22:6246–6253
Henry T, Iwen PC, Hinrichs SH (2000) Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J Clin Microbiol 38:1510–1515
IARC (2002) Some traditional herbal medicines, some mycotoxins, naphthalene and styrene. IARC Monogr Eval Carcinog Risks Hum 82:1–556
Inan F, Pala M, Doymaz I (2007) Use of ozone in detoxification of aflatoxin B1 in red pepper. J Stored Prod Res 43:425–429
Jiang J, Yan L, Ma Z (2009) Molecular characterization of an atoxigenic Aspergillus flavus strain AF051. Appl Microbiol Biotechnol 83(3):501–505
ISTA (2005) International rules for seed testing edition Bassersdorf CH–Switzerland. International Seed Testing Association
Kabak B (2009) The fate of mycotoxins during thermal food processing. J Sci Food Agric 89:549–554
Kensler TW, Roebuck BD, Wogan GN, Groopman JD (2010) Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicol Sci 120:S28–S48
Kim DM, Chung SH, Chun HS (2011) Multiplex PCR assay for the detection of aflatoxigenic and non-aflatoxigenic fungi in meju, a Korean fermented soybean food starter. Food Microbiol 28:1402–1408
Klich MA (2002) Identification of common Aspergillus species. Centraalbureau voor Schimmelcultures, Utrecht
Klich MA (2007) Aspergillus flavus: the major producer of aflatoxin. Mol Plant Pathol 8:713–722
Kumar S, Tamura K, Nei M (1994) MEGA: Molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci CABIOS 10(2):189–191
Kurtzman CP, Horn BW, Hesseltine CW (1987) Aspergillus nomius, a new aflatoxin-producing species related to Aspergillus flavus and Aspergillus tamarii. Antonie Van Leeuwenhoek 53:147–158
Lin MT, Dianese JC (1976) A coconut-agar medium for rapid detection of aflatoxin production by Aspergillus spp. Phytopathology 66:1466–1469
Logotheti M, Kotsovili-Tseleni A, Arsenis G, Legakis NI (2009) Multiplex PCR for the discrimination of A. fumigatus, A. flavus, A. niger and A. terreus. J Microbiol Methods 76:209–211
Mayer Z, Bagnara A, Färber P, Geisen R (2003) Quantification of the copy number of nor-1, a gene of the aflatoxin biosynthetic pathway by real-time PCR, and its correlation to the cfu of Aspergillus flavus in foods. Int J Food Microbiol 82:143–151
Mishra HN, Das C (2003) A review on biological control and metabolism of aflatoxin. Crit Rev Food Sci Nutr 43(3):245–264
Muscarella M, Iammarino M, Nardiello D, Lo Magro S, Palermo C, Centonze D, Palermo D (2009) Validation of a confirmatory analytical method for the determination of aflatoxins B1, B2, G1 and G2 in foods and feed materials by HPLC with on-line photochemical derivatization and fluorescence detection. Food Addit Contam 26:1402–1410
Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson K-H (2008) Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinforma 4:653
Novas MV, Cabral D (2002) Association of mycotoxin and sclerotia production with compatibility groups in Aspergillus flavus from peanut in Argentina. Plant Dis 86:215–219
Pitt JI, Hocking AD, Glenn DR (1983) An improved medium for the detection of Aspergillus flavus and A. parasiticus. J Appl Bacteriol 54(1):109–114
Rodrigues P, Venâncio A, Kozakiewicz Z, Lima N (2009) A polyphasic approach to the identification of aflatoxigenic and non-aflatoxigenic strains of Aspergillus section Flavi isolated from Portuguese almonds. Int J Food Microbiol 129:187–193
Saito M, Machida S (1999) A rapid identification method for aflatoxin-producing strains of Aspergillus flavus and A. parasiticus by ammonia vapor. Mycoscience 40:205–208
Scherm B, Palomba M, Serra D, Marcello A, Migheli Q (2005) Detection of transcripts of the aflatoxin genes aflD, aflO, and aflP by reverse transcription–polymerase chain reaction allows differentiation of aflatoxin-producing and non-producing isolates of Aspergillus flavus and Aspergillus parasiticus. Int J Food Microbiol 98:201–210
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Shapira R, Paster N, Eyal O, Menasherov M, Mett A, Salomon R (1996) Detection of aflatoxigenic molds in grains by PCR. Appl Environ Microbiol 62:3270–3273
Shetty PH, Hald B, Jespersen L (2007) Surface binding of aflatoxin B1 by Saccharomyces cerevisiae strains with potential decontaminating abilities in indigenous fermented foods. Int J Food Microbiol 113(1):41–46
Shier WT, Lao Y, Steele TW, Abbas HK (2005) Yellow pigments used in rapid identification of aflatoxin-producing Aspergillus strains are anthraquinones associated with the aflatoxin biosynthetic pathway. Bioorganic Chem 33:426–438
Singh K (1991) An illustrated manual on identification of some seed-borne Aspergilli, Fusaria, Penicillia and their mycotoxins. Danish Government Institute of Seed Pathology for Developing Countries, Copenhagen
Sweeney MJ, Pàmies P, Dobson AD (2000) The use of reverse transcription-polymerase chain reaction (RT-PCR) for monitoring aflatoxin production in Aspergillus parasiticus 439. Int J Food Microbiol 56:97–103
Tran-Dinh N, Pitt JI, Carter DA (1999) Molecular genotype analysis of natural toxigenic and nontoxigenic isolates of Aspergillus flavus and A. parasiticus. Mycol Res 103:1485–1490
Tran-Dinh N, Pitt JI, Markwell PJ (2014) Selection of non-toxigenic strains of Aspergillus flavus for biocontrol of aflatoxins in maize in Thailand. Biocontrol Sci. Technol. 24:652–661
Trucksess MW (2000) Natural toxins. In: Horwitz W (ed) Official methods of analysis of AOAC International, 17th edn. AOAC International, Gaithersburg
Vipin AV, Rao R, Kurrey NK, Anu Appaiah KA, Venkateswaran G (2017) Protective effects of phenolics rich extract of ginger against aflatoxin B1-induced oxidative stress and hepatotoxicity. Biomed Pharmacother 91:415–424
Wei D, Zhou L, Selvaraj JN, Zhang C, Xing F, Zhao Y, Wang Y, Liu Y (2014) Molecular characterization of atoxigenic Aspergillus flavus isolates collected in China. J Microbiol 52:559–565
White TJ, Bruns T, Lee S, Taylor JW et al (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc Guide Methods Appl 18:315–322
Yin Y, Lou T, Yan L, Michailides TJ, Ma Z (2009) Molecular characterization of toxigenic and atoxigenic Aspergillus flavus isolates, collected from peanut fields in China. J Appl Microbiol 107:1857–1865
Yu J, Chang P-K, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, Payne GA, Linz JE, Woloshuk CP, Bennett JW (2004) Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol 70:1253–1262
Acknowledgements
The authors are thankful to Director, CSIR- Central Food Technological Research Institute, for providing necessary facilities to carry out this study and CSIR for the financial support in the form of Junior and senior research fellowship to the first author. Authors would also like to thank Dr. Anu Appaiah K. A., Principal scientist, Microbiology and Fermentation Technology, CSIR-CFTRI, for his kind support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
203_2020_1822_MOESM1_ESM.docx
Fig.S1. Screening and isolation Aspergillus flavus from soil and seed samples. a. Screening through standard plate method; b. Screening through standard blotter method; c.Aspergillus flavus on PDA; d. Microscopic view of A. flavus; e. Culture methods to differentiate aflatoxigenic and non-aflatoxigenic A. flavusi. Ultraviolet test; ii. Ammonia vapor test; iii. Yellow pigmentation (DOCX 1015 kb)
Rights and permissions
About this article
Cite this article
Rao, K.R., Vipin, A.V. & Venkateswaran, G. Molecular profile of non-aflatoxigenic phenotype in native strains of Aspergillus flavus. Arch Microbiol 202, 1143–1155 (2020). https://doi.org/10.1007/s00203-020-01822-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-01822-1