Abstract
Purpose
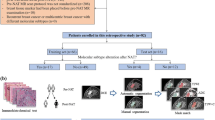
Volumetric assessment of meningiomas represents a valuable tool for treatment planning and evaluation of tumor growth as it enables a more precise assessment of tumor size than conventional diameter methods. This study established a dedicated meningioma deep learning model based on routine magnetic resonance imaging (MRI) data and evaluated its performance for automated tumor segmentation.
Methods
The MRI datasets included T1-weighted/T2-weighted, T1-weighted contrast-enhanced (T1CE) and FLAIR of 126 patients with intracranial meningiomas (grade I: 97, grade II: 29). For automated segmentation, an established deep learning model architecture (3D deep convolutional neural network, DeepMedic, BioMedIA) operating on all four MR sequences was used. Segmentation included the following two components: (i) contrast-enhancing tumor volume in T1CE and (ii) total lesion volume (union of lesion volume in T1CE and FLAIR, including solid tumor parts and surrounding edema). Preprocessing of imaging data included registration, skull stripping, resampling, and normalization. After training of the deep learning model using manual segmentations by 2 independent readers from 70 patients (training group), the algorithm was evaluated on 56 patients (validation group) by comparing automated to ground truth manual segmentations, which were performed by 2 experienced readers in consensus.
Results
Of the 56 meningiomas in the validation group 55 were detected by the deep learning model. In these patients the comparison of the deep learning model and manual segmentations revealed average dice coefficients of 0.91 ± 0.08 for contrast-enhancing tumor volume and 0.82 ± 0.12 for total lesion volume. In the training group, interreader variabilities of the 2 manual readers were 0.92 ± 0.07 for contrast-enhancing tumor and 0.88 ± 0.05 for total lesion volume.
Conclusion
Deep learning-based automated segmentation yielded high segmentation accuracy, comparable to manual interreader variability.




Similar content being viewed by others
Abbreviations
- FLAIR:
-
Fluid-attenuated inversion recovery
- T1CE:
-
T1-weighted gadolinium contrast enhanced
- WHO:
-
World Health Organization
References
DeAngelis LM. Brain tumors. N Engl J Med. 2001;344:114–23.
Schob S, Frydrychowicz C, Gawlitza M, Bure L, Preuß M, Hoffmann KT, Surov A. Signal intensities in preoperative MRI do not reflect proliferative activity in meningioma. Transl Oncol. 2016;9:274–9.
Vernooij MW, Ikram MA, Tanghe HL, Vincent AJ, Hofman A, Krestin GP, Niessen WJ, Breteler MM, van der Lugt A. Incidental findings on brain MRI in the general population. N Engl J Med. 2007;357:1821–8.
Spasic M, Pelargos PE, Barnette N, Bhatt NS, Lee SJ, Ung N, Gopen Q, Yang I. Incidental meningiomas. Neurosurg Clin N Am. 2016;27:229–38.
Yao A, Pain M, Balchandani P, Shrivastava RK. Can MRI predict meningioma consistency?: a correlation with tumor pathology and systematic review. Neurosurg Rev. 2018;41:745–53.
Goldbrunner R, Minniti G, Preusser M, Jenkinson MD, Sallabanda K, Houdart E, von Deimling A, Stavrinou P, Lefranc F, Lund-Johansen M, Moyal EC, Brandsma D, Henriksson R, Soffietti R, Weller M. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol. 2016;17:e383–91.
Chang V, Narang J, Schultz L, Issawi A, Jain R, Rock J, Rosenblum M. Computer-aided volumetric analysis as a sensitive tool for the management of incidental meningiomas. Acta Neurochir (Wien). 2012;154:589–97.
Fountain DM, Soon WC, Matys T, Guilfoyle MR, Kirollos R, Santarius T. Volumetric growth rates of meningioma and its correlation with histological diagnosis and clinical outcome: a systematic review. Acta Neurochir (Wien). 2017;159:435–45.
Zeidman LA, Ankenbrandt WJ, Du H, Paleologos N, Vick NA. Growth rate of non-operated meningiomas. J Neurol. 2008;255:891–5.
Xue Y, Chen S, Qin J, Liu Y, Huang B, Chen H. Application of deep learning in automated analysis of molecular images in cancer: a survey. Contrast Media Mol Imaging. 2017;2017:9512370.
Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ. Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging. 2017;30:449–59.
Mazzara GP, Velthuizen RP, Pearlman JL, Greenberg HM, Wagner H. Brain tumor target volume determination for radiation treatment planning through automated MRI segmentation. Int J Radiat Oncol Biol Phys. 2004;59:300–12.
Lotan E, Jain R, Razavian N, Fatterpekar GM, Lui YW. State of the art: machine learning applications in glioma imaging. AJR Am J Roentgenol. 2019;212:26–37.
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Ç, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SM, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging. 2015;34:1993–2024.
Baessler B, Mannil M, Oebel S, Maintz D, Alkadhi H, Manka R. Subacute and chronic left ventricular myocardial scar: accuracy of texture analysis on nonenhanced cine MR images. Radiology. 2018;286:103–12.
Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2016;36:61–78.
Laukamp KR, Thiele F, Shakirin G, Zopfs D, Faymonville A, Timmer M, Maintz D, Perkuhn M, Borggrefe J. Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur Radiol. 2019;29:124–32.
Perkuhn M, Stavrinou P, Thiele F, Shakirin G, Mohan M, Garmpis D, Kabbasch C, Borggrefe J. Clinical evaluation of a multiparametric deep learning model for glioblastoma segmentation using heterogeneous magnetic resonance imaging data from clinical routine. Invest Radiol. 2018;53:647–54.
Kickingereder P, Isensee F, Tursunova I, Petersen J, Neuberger U, Bonekamp D, Brugnara G, Schell M, Kessler T, Foltyn M, Harting I, Sahm F, Prager M, Nowosielski M, Wick A, Nolden M, Radbruch A, Debus J, Schlemmer HP, Heiland S, Platten M, von Deimling A, van den Bent MJ, Gorlia T, Wick W, Bendszus M, Maier-Hein KH. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 2019;20:728–40.
Kooi T, Litjens G, van Ginneken B, Gubern-Mérida A, Sánchez CI, Mann R, den Heeten A, Karssemeijer N. Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal. 2017;35:303–12.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44.
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–20.
Lindemann F, Laukamp KR, Jacobs A, Hinrichs KH. Interactive comparative visualization of multimodal brain tumor segmentation data. Vision, modeling, and visualization. 2013. pp. 105–12.
GitHub. DeepMedic. 2019. https://github.com/deepmedic. Accessed 21 Nov 2019.
Crum WR, Camara O, Hill DLG. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans Med Imaging. 2006;25:1451–61.
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31.
Hoseini F, Shahbahrami A, Bayat P. An efficient implementation of deep convolutional neural networks for MRI segmentation. J Digit Imaging. 2018;31:738–47.
Shimol EB, Joskowicz L, Eliahou R, Shoshan Y. Computer-based radiological longitudinal evaluation of meningiomas following stereotactic radiosurgery. Int J Comput Assist Radiol Surg. 2018;13:215–28.
Laukamp KR, Lindemann F, Weckesser M, Hesselmann V, Ligges S, Wölfer J, Jeibmann A, Zinnhardt B, Viel T, Schäfers M, Paulus W, Stummer W, Schober O, Jacobs AH. Multimodal imaging of patients with gliomas confirms (11)C-MET PET as a complementary marker to MRI for noninvasive tumor grading and intraindividual follow-up after therapy. Mol Imaging. 2017;16:1536012116687651.
Sauwen N, Acou M, Bharath HN, Sima DM, Veraart J, Maes F, Himmelreich U, Achten E, Van Huffel S. The successive projection algorithm as an initialization method for brain tumor segmentation using non-negative matrix factorization. PLoS One. 2017;12:e180268.
Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature. 2013;495:187–92.
Badve C, Yu A, Dastmalchian S, Rogers M, Ma D, Jiang Y, Margevicius S, Pahwa S, Lu Z, Schluchter M, Sunshine J, Griswold M, Sloan A, Gulani V. MR fingerprinting of adult brain tumors: initial experience. AJNR Am J Neuroradiol. 2017;38:492–9.
Laukamp KR, Shakirin G, Baeßler B, Thiele F, Zopfs D, Hokamp NG, Timmer M, Kabbasch C, Perkuhn M, Borggrefe J. Accuracy of radiomics-based feature analysis on multiparametric MR images for non-invasive meningioma grading. World Neurosurg. 2019;132:e366–90.
Latini F, Larsson EM, Ryttlefors M. Rapid and accurate MRI segmentation of peritumoral brain edema in meningiomas. Clin Neuroradiol. 2017;27:145–52.
Otero-Rodriguez A, Tabernero MD, Munoz-Martin MC, Sousa P, Orfao A, Pascual-Argente D, Gonzalez-Tablas M, Ruiz-Martin L. Re-evaluating Simpson grade I, II, and III resections in neurosurgical treatment of world health organization grade I meningiomas. World Neurosurg. 2016;96:483–8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
G. Shakirin, F. Thiele, M. Perkuhn are Philips employees. J. Borggrefe received an honorarium from Philips for scientific lectures. K.R. Laukamp, L. Pennig, R. Reimer, L. Goertz, D. Zopfs and M. Timmer declare that they have no competing interests.
Additional information
Patient population
Of the 126 patients included in this study 56 have been analyzed previously by this group and published in 2018 in European Radiology, where they also served as segmentation ground truth. To improve comparability between the deep learning models used in this study and in order to determine whether retraining by a specific tumor type is warranted, the former cohort was used again as segmentation ground truth.
Rights and permissions
About this article
Cite this article
Laukamp, K.R., Pennig, L., Thiele, F. et al. Automated Meningioma Segmentation in Multiparametric MRI. Clin Neuroradiol 31, 357–366 (2021). https://doi.org/10.1007/s00062-020-00884-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00062-020-00884-4