Abstract
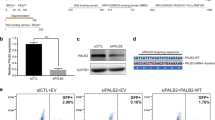
The PALB2 protein is essential to RAD51-mediated homologous recombination (HR) repair. Germline monoallelic PALB2 pathogenic variants confer significant risks for breast cancer. However, the majority of PALB2 variants remain classified as variants of unknown significance (VUS). We aim to functionally and mechanistically evaluate three novel PALB2 VUS. Patient-derived lymphoblastoid cell lines containing the VUS were analyzed for nuclear localization and foci formation of RAD51 as a measure of HR efficiency. To understand the mechanism underlying the HR deficiency, PALB2 nuclear localization was assessed using immunofluorescence studies. Among these VUS, c.3251C>T (p.Ser1084Leu) occurred in a patient with metastatic breast cancer while c.1054G>C (p.Glu352Gln) and c.1057A>G (p.Lys353Glu) were seen in patients with squamous cell carcinoma of skin and renal cell carcinoma respectively. Variant c.3251C>T was located within the WD40 domain which normally masked the nuclear export signal sequence responsible for nuclear delocalization of PALB2. Correspondingly, c.3251C>T displayed aberrant cytoplasmic localization of PALB2 which led to an impaired RAD51 nuclear localization and foci formation. On the other hand, both c.1054G>C and c.1057A>G showed intact HR functions and nuclear localization of PALB2, consistent with their locations within domains of no known function. Additionally, the prevalence of c.1054G>C was similar among healthy controls and patients with breast cancer (as seen in other studies), suggestive of its non-pathogenicity. In conclusion, our studies provided the functional evidence showing the deleterious effect of c.3251C>T, and non-deleterious effects of c.1054G>C and c.1057A>G. Using the ClinGen Pathogenicity calculator, c.3251C>T remains a VUS while c.1054G>C and c.1057A>G may be classified as likely benign variants.



Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Buisson R, Masson J-Y (2012) PALB2 self-interaction controls homologous recombination. Nucl Acids Res 40(20):10312–10323
Park J-Y, Singh TR, Nassar N, Zhang F, Freund M, Hanenberg H, Meetei AR, Andreassen PR (2014) Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene 33(40):4803–4812
Ducy M, Sesma-Sanz L, Guitton-Sert L, Lashgari A, Gao Y, Brahiti N, Rodrigue A, Margaillan G, Caron M-C, Côté J (2019) The tumor suppressor PALB2: inside out. Trends Biochem Sci 44(3):226–240
Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J (2007) Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet 39(2):159–161
Yang X, Leslie G, Doroszuk A, Schneider S, Allen J, Decker B, Dunning AM, Redman J, Scarth J, Plaskocinska I, Luccarini C, Shah M, Pooley K, Dorling L, Lee A, Adank MA, Adlard J, Aittomäki K, Andrulis IL, Ang P, Barwell J, Bernstein JL, Bobolis K, Borg Å, Blomqvist C, Claes KBM, Concannon P, Cuggia A, Culver JO, Damiola F, de Pauw A, Diez O, Dolinsky JS, Domchek SM, Engel C, Evans DG, Fostira F, Garber J, Golmard L, Goode EL, Gruber SB, Hahnen E, Hake C, Heikkinen T, Hurley JE, Janavicius R, Kleibl Z, Kleiblova P, Konstantopoulou I, Kvist A, Laduca H, Lee ASG, Lesueur F, Maher ER, Mannermaa A, Manoukian S, McFarland R, McKinnon W, Meindl A, Metcalfe K, Mohd Taib NA, Moilanen J, Nathanson KL, Neuhausen S, Ng PS, Nguyen-Dumont T, Nielsen SM, Obermair F, Offit K, Olopade OI, Ottini L, Penkert J, Pylkäs K, Radice P, Ramus SJ, Rudaitis V, Side L, Silva-Smith R, Silvestri V, Skytte A-B, Slavin T, Soukupova J, Tondini C, Trainer AH, Unzeitig G, Usha L, van Overeem HT, Whitworth J, Wood M, Yip CH, Yoon S-Y, Yussuf A, Zogopoulos G, Goldgar D, Hopper JL, Chenevix-Trench G, Pharoah P, George SHL, Balmaña J, Houdayer C, James P, El-Haffaf Z, Ehrencrona H, Janatova M, Peterlongo P, Nevanlinna H, Schmutzler R, Teo S-H, Robson M, Pal T, Couch F, Weitzel JN, Elliott A, Southey M, Winqvist R, Easton DF, Foulkes WD, Antoniou AC, Tischkowitz M (2019) Cancer risks associated with germline PALB2 pathogenic variants: an international study of 524 families. J Clin Oncol 12:56–89
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips K-A, Mooij TM, Roos-Blom M-J, Jervis S, Van Leeuwen FE, Milne RL, Andrieu N (2017) Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. Jama 317(23):2402–2416
Cybulski C, Kluźniak W, Huzarski T, Wokołorczyk D, Kashyap A, Jakubowska A, Szwiec M, Byrski T, Dębniak T, Górski B (2015) Clinical outcomes in women with breast cancer and a PALB2 mutation: a prospective cohort analysis. Lancet Oncol 16(6):638–644
Desmond A, Kurian AW, Gabree M, Mills MA, Anderson MJ, Kobayashi Y, Horick N, Yang S, Shannon KM, Tung N (2015) Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment. JAMA Oncol 1(7):943–951
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody W, Hegde M, Lyon E, Spector E (2015) ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424
Caleca L, Catucci I, Figlioli G, De Cecco L, Pesaran T, Ward M, Volorio S, Falanga A, Marchetti M, Iascone M (2018) Missense variants detected in breast cancer families preventing BRCA2-PALB2 protein interaction. Front Oncol 8:480
Phuah SY, Lee SY, Kang P, Kang IN, Yoon S-Y, Thong MK, Hartman M, Sng J-H, Yip CH, Taib NAM, Teo S-H (2013) Prevalence of PALB2 mutations in breast cancer patients in multi-ethnic Asian population in Malaysia and Singapore. PLoS ONE 8(8):e73638
Piffer A, Luporsi E, Mathelin C (2018) PALB2, gène majeur de susceptibilité au cancer du sein. Gynécologie Obstétrique Fertilité Sénologie 46(10):701–705
NCBI ClinVar. https://www.ncbi.nlm.nih.gov/clinvar/?term=palb2%5Bgene%5D. Last Accessed Dec 2019
Tischkowitz M, Xia B, Sabbaghian N, Reis-Filho JS, Hamel N, Li G, van Beers EH, Li L, Khalil T, Quenneville LA (2007) Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci USA 104(16):6788–6793
Hanenberg H, Andreassen PR (2018) PALB2 (partner and localizer of BRCA2). Atlas Genet Cytogenet Oncol Haematol 22(12):484–490
Wong-Brown MW, Avery‐Kiejda KA, Bowden NA, Scott RJ (2014) Low prevalence of germline PALB2 mutations in Australian triple‐negative breast cancer. Int J Cancer 134(2):301–305
Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkäs K, Roberts J, Lee A, Subramanian D, De Leeneer K, Fostira F, Tomiak E, Neuhausen SL, Teo ZL, Khan S, Aittomäki K, Moilanen JS, Turnbull C, Seal S, Mannermaa A, Kallioniemi A, Lindeman GJ, Buys SS, Andrulis IL, Radice P, Tondini C, Manoukian S, Toland AE, Miron P, Weitzel JN, Domchek SM, Poppe B, Claes KBM, Yannoukakos D, Concannon P, Bernstein JL, James PA, Easton DF, Goldgar DE, Hopper JL, Rahman N, Peterlongo P, Nevanlinna H, King M-C, Couch FJ, Southey MC, Winqvist R, Foulkes WD, Tischkowitz M (2014) Breast-cancer risk in families with mutations in PALB2. N Engl J Med 371(6):497–506
Heikkinen T, Kärkkäinen H, Aaltonen K, Milne RL, Heikkilä P, Aittomäki K, Blomqvist C, Nevanlinna H (2009) The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin Cancer Res 15(9):3214–3222
Pauty J, Couturier AM, Rodrigue A, Caron M-C, Coulombe Y, Dellaire G, Masson J-Y (2017) Cancer-causing mutations in the tumor suppressor PALB2 reveal a novel cancer mechanism using a hidden nuclear export signal in the WD40 repeat motif. Nucl Acids Res 45(5):2644–2657
Amélie Rodrigue GM, Gomes TT, Coulombe Y, Ducy M, da Costa e Silva Carvalho S, De-Gregoriis G, de Souza LM, Dellaire G, da Silva Junior WA, Monteiro A, Carvalho M, Simard J, Masson J-Y (2019) A global functional analysis of missense mutations reveals two major hotspots in the PALB2 tumor suppressor. Nucleic Acids Res 47(20):10662–10677
Tchernitchko D, Goossens M, Wajcman H (2004) In silico prediction of the deleterious effect of a mutation: proceed with caution in clinical genetics. Clin Chem 50(11):1974–1978
Bleuyard JY, Buisson R, Masson JY, Esashi F (2012) ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep 13(2):135–141
Erkko H, Xia B, Nikkilä J, Schleutker J, Syrjäkoski K, Mannermaa A, Kallioniemi A, Pylkäs K, Karppinen S-M, Rapakko K, Miron A, Sheng Q, Li G, Mattila H, Bell DW, Haber DA, Grip M, Reiman M, Jukkola-Vuorinen A, Mustonen A, Kere J, Aaltonen LA, Kosma V-M, Kataja V, Soini Y, Drapkin RI, Livingston DM, Winqvist R (2007) A recurrent mutation in PALB2 in Finnish cancer families. Nature 446:316–319
MacArthur D, Manolio T, Dimmock D, Rehm H, Shendure J, Abecasis G, Adams D, Altman R, Antonarakis S, Ashley E (2014) Guidelines for investigating causality of sequence variants in human disease. Nature 508(7497):469–476
Patel RY, Shah N, Jackson AR, Ghosh R, Pawliczek P, Paithankar S, Baker A, Riehle K, Chen H, Milosavljevic S, Bizon C, Rynearson S, Nelson T, Jarvik GP, Rehm HL, Harrison SM, Azzariti D, Powell B, Babb L, Plon SE, Milosavljevic A, on behalf of the ClinGen R (2017) ClinGen pathogenicity calculator: a configurable system for assessing pathogenicity of genetic variants. Genome Med 9(1):3
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Neale BM, Daly MJ, MacArthur DG (2019) Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv.https://doi.org/10.1101/531210
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4(7):1073–1081
Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A (2006) Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet 43(4):295–305
Toh MR, Chiang JB, Chong ST, Chan SH, Ishak NDB, Courtney E, Lee WH, Syed Abdillah Al SMFB, Carson Allen J, Jr Lim KH (2018) Germline pathogenic variants in homologous recombination and DNA repair genes in an Asian cohort of young-onset colorectal cancer. JNCI Cancer Spectrum 2(4):054
Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K (2010) REAP: a two minute cell fractionation method. BMC Res Notes 3:294
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671
Acknowledgements
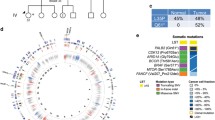
We would like to thank the patients and research participants for their contribution to the study. We would like to acknowledge St Jude PeCan Data Portal for the web application in generating Fig. 1e.
Funding
We thank our sources of support: National Medical Research Council (CSA) (NMRC/CSA-INV/0017/2017) and Singhealth Foundation Research Grant (SHF/PRISM002/2015) to JN and SingHealth (SMSTDA-Project FY2018) to MR. JYM is a FRQS chair in genome stability and was supported by a CIHR Foundation grant.
Author information
Authors and Affiliations
Contributions
CE, MR, ST, SH, ND, EC and AMK made substantial contributions to the design, administration and conceptualization of the study, interpretation of data and critically revising and drafting the manuscript. AR and JYM provided expertise and reagents. JN provided funding, acquisition of data, conception and design of the study, manuscript review and editing. All authors approved the submission of the final manuscript and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the Singhealth Centralized Institutional Review Board (IRB 2010/426/B) with signed inform consent from participants.
Rights and permissions
About this article
Cite this article
Toh, M.R., Low, C.E., Chong, S.T. et al. Missense PALB2 germline variant disrupts nuclear localization of PALB2 in a patient with breast cancer. Familial Cancer 19, 123–131 (2020). https://doi.org/10.1007/s10689-020-00163-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-020-00163-8