Abstract
Introduction
Bioengineering an implantable artificial kidney (IAK) will require renal epithelial cells capable of reabsorption of salt and water. We used genome engineering to modify cells for improved Na+/H+ exchange and H2O reabsorption. The non-viral piggyBac transposon system enables genome engineering cells to stably overexpress one or more transgenes simultaneously.
Methods
We generated epitope-tagged human sodium hydrogen exchanger 3 (NHE3) and aquaporin-1 (AQP1) cDNA expressing piggyBac transposon vectors. Transgene expression was evaluated via western blot and immunofluorescence. Flow cytometry analysis was used to quantitate transporter expression in a library of genome engineered clones. Cell surface biotinylation was used evaluate surface protein localization. Blister formation assays were used to monitor cellular volumetric transport.
Results
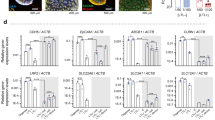
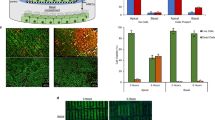
piggyBac enabled stable transposon integration and overexpression of cumate-inducible NHE3 and/or constitutively expressing AQP1 in cultured renal (MDCK) epithelial cells. Cell surface delivery of NHE3 and AQP1 was confirmed using cell surface biotinylation assays. Flow cytometry of a library of MDCK clones revealed varying expression of AQP1 and NHE3. MDCK cells expressing AQP1 and cumate-inducible NHE3 demonstrated increased volumetric transport.
Conclusions
Our results demonstrate that renal epithelial cells an be genome engineered for enhanced volumetric transport that will be needed for an IAK device. Our results lay the foundation for future studies of genome engineering human kidney cells for renal tubule cell therapy.





Similar content being viewed by others
References
Brant, S. R., M. Bernstein, J. J. Wasmuth, E. W. Taylor, J. D. McPherson, X. Li, et al. Physical and genetic mapping of a human apical epithelial Na +/H + exchanger (NHE3) isoform to chromosome 5p15.3. Genomics 15(3):668–672, 1993.
Cary, L. C., M. Goebel, B. G. Corsaro, H. G. Wang, E. Rosen, and M. J. Fraser. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 172(1):156–169, 1989.
Cattaneo, I., L. Condorelli, A. R. Terrinoni, L. Antiga, F. Sangalli, and A. Remuzzi. Shear stress reverses dome formation in confluent renal tubular cells. Cell. Physiol. Biochem. 28(4):673–682, 2011.
Doherty, J. E., L. E. Huye, K. Yusa, L. Zhou, N. L. Craig, and M. H. Wilson. Hyperactive piggyBac gene transfer in human cells and in vivo. Hum. Gene Ther. 23(3):311–320, 2012.
Doherty, J. E., L. E. Woodard, A. S. Bear, A. E. Foster, and M. H. Wilson. An adaptable system for improving transposon-based gene expression in vivo via transient transgene repression. FASEB J. 27(9):3753–3762, 2013.
Elick, T. A., C. A. Bauser, and M. J. Fraser. Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica. 98(1):33–41, 1996.
Fraser, M. J., T. Ciszczon, T. Elick, and C. Bauser. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect MolBiol. 5(2):141–151, 1996.
Gaush, C. R., W. L. Hard, and T. F. Smith. Characterization of an established line of canine kidney cells (MDCK). Proc. Soc. Exp. Biol. Med. Soc. Exp. Biol. Med (N.Y.). 122(3):931–935, 1966.
Hallman, M. A., S. Zhuang, and R. G. Schnellmann. Regulation of dedifferentiation and redifferentiation in renal proximal tubular cells by the epidermal growth factor receptor. J. Pharmacol. Exp. Ther. 325(2):520–528, 2008.
Hew, B. E., R. Sato, D. Mauro, I. Stoytchev, and J. B. Owens. RNA-guided piggyBac transposition in human cells. Synth Biol (Oxf). 4(1):ysz018, 2019.
Kahlig, K. M., S. K. Saridey, A. Kaja, M. A. Daniels, A. L. George, Jr, and M. H. Wilson. Multiplexed transposon-mediated stable gene transfer in human cells. Proc. Natl. Acad. Sci. USA 107(4):1343–1348, 2010.
Klebe, R. J., A. Grant, G. Grant, and P. Ghosh. Cyclic-AMP deficient MDCK cells form tubules. J. Cell. Biochem. 59(4):453–462, 1995.
Lever, J. E. Regulation of dome formation in differentiated epithelial cell cultures. J. Supramol. Struct. 12(2):259–272, 1979.
Love, H. D., M. Ao, S. Jorgensen, L. Swearingen, N. Ferrell, R. Evans, et al. Substrate elasticity governs differentiation of renal tubule cells in prolonged culture. Tissue Eng. Part A. 25(13–14):1013–1022, 2019.
Luo, W., D. L. Galvan, L. E. Woodard, D. Dorset, S. Levy, and M. H. Wilson. Comparative analysis of chimeric ZFP-, TALE- and Cas9-piggyBac transposases for integration into a single locus in human cells. Nucleic Acids Res. 45(14):8411–8422, 2017.
McCullough, K. P., H. Morgenstern, R. Saran, W. H. Herman, and B. M. Robinson. Projecting ESRD incidence and prevalence in the united states through 2030. J. Am. Soc. Nephrol. 30(1):127–135, 2019.
Mullick, A., Y. Xu, R. Warren, M. Koutroumanis, C. Guilbault, S. Broussau, et al. The cumate gene-switch: a system for regulated expression in mammalian cells. BMC Biotechnol. 6:43, 2006.
Nielsen, S., B. L. Smith, E. I. Christensen, M. A. Knepper, and P. Agre. CHIP28 water channels are localized in constitutively water-permeable segments of the nephron. J. Cell Biol. 120(2):371–383, 1993.
Perez-Pinera, P., D. D. Kocak, C. M. Vockley, A. F. Adler, A. M. Kabadi, L. R. Polstein, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 10(10):973–976, 2013.
Saha, S., L. E. Woodard, E. M. Charron, R. C. Welch, C. M. Rooney, and M. H. Wilson. Evaluating the potential for undesired genomic effects of the piggyBac transposon system in human cells. Nucleic Acids Res. 43(3):1770–1782, 2015.
Saito, S., Y. Nakazawa, A. Sueki, K. Matsuda, M. Tanaka, R. Yanagisawa, et al. Anti-leukemic potency of piggyBac-mediated CD19-specific T cells against refractory Philadelphia chromosome-positive acute lymphoblastic leukemia. Cytotherapy. 16(9):1257–1269, 2014.
Salani, M., S. Roy, and W. H. T. Fissell. Innovations in wearable and implantable artificial kidneys. Am J Kidney Dis. 72(5):745–751, 2018.
Saridey, S. K., L. Liu, J. E. Doherty, A. Kaja, D. L. Galvan, B. S. Fletcher, et al. PiggyBac transposon-based inducible gene expression in vivo after somatic cell gene transfer. Mol. Ther. 17(12):2115–2120, 2009.
Su, H. W., H. H. Yeh, S. W. Wang, M. R. Shen, T. L. Chen, P. R. Kiela, et al. Cell confluence-induced activation of signal transducer and activator of transcription-3 (Stat3) triggers epithelial dome formation via augmentation of sodium hydrogen exchanger-3 (NHE3) expression. J. Biol. Chem. 282(13):9883–9894, 2007.
Wang, W., C. Lin, D. Lu, Z. Ning, T. Cox, D. Melvin, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 105(27):9290–9295, 2008.
Wilson, M., and L. Limbird. Mechanisms regulating the cell surface residence time of the alpha(2A)-adrenergic receptor. Biochemistry. 39(4):693–700, 2000.
Woodard, L. E., and M. H. Wilson. piggyBac-ing models and new therapeutic strategies. Trends Biotechnol. 33(9):525–533, 2015.
Acknowledgments
These studies were supported by DK093360 (NIH) and BX004285 (VA) to MHW, EB021214 to SR and WHF, and the Vanderbilt O’Brien Kidney Center Cell and Genome Engineering Core (DK114809). The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). Brittany K. Matlock in the VMC Flow Cytometry Shared Resource provided valuable assistance in developing the intracellular staining protocol for high-throughput sampling. Confocal immunofluorescence image collection and data analysis were performed in part through the use of the VUCell Imaging Shared Resource (supported by NIHgrants CA68485, DK20593, DK58404, DK59637 and EY08126) and NIH S10 Grant Number 1S10RR027396-01. Mouse anti-myc antibody was produced by the Vanderbilt Antibody and Protein Resource. The Vanderbilt Antibody and Protein Resource is supported by the Vanderbilt Institute of Chemical Biology and the Vanderbilt Ingram Cancer Center (P30 CA68485).
Conflict of interest
MHW, RAV, WL, and RCW declare no conflicts of interest. SR and WHF are founders of Silicon Kidney.
Ethical Standards
No human or animal studies were carried out by the authors for this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Michael R. King oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wilson, M.H., Veach, R.A., Luo, W. et al. Genome Engineering Renal Epithelial Cells for Enhanced Volume Transport Function. Cel. Mol. Bioeng. 13, 17–26 (2020). https://doi.org/10.1007/s12195-019-00601-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-019-00601-3