Abstract
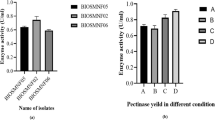
Pectinolytic enzymes that catalyze the breakdown of substrates containing pectin are widespread. Pectinases have potential applications in various industries, including food, animal feed, textile, paper, and fuel. In this study, one hundred bacterial isolates were collected from Marand city farmlands (Azarbaijan-E-Sharqi, Iran) and screened by MP medium on the base of pectinase activity considering the significance of pectinases. The results depicted that three isolates showed the most pectinase activity (more massive halo). The biochemical and molecular test results showed that the three screened bacteria were Enterobacter and named Enterobacter sp. MF41, Enterobacter sp. MF84, and Enterobacter sp. MF90. Enterobacter sp. MF84 had the largest halo, so this strain was selected for the study of its produced pectinase. The results exhibited that the produced enzyme has optimum temperature and pH for activity at 30 °C and in 9, respectively. Finally, the enzyme production by Enterobacter sp. MF84 is optimized using response surface methodology (RSM) considering four factors (NH4Cl, K2HPO4, pectin, and incubation time) as variables. The results showed that the optimization procedure increased the enzyme production up to 12 times (from 1.16 to 14.16 U/mg). The Pareto analysis revealed that ammonium chloride has a significant role in decreasing the enzyme production, probably by inducing the nitrification pathway enzymes in the presence of organic nitrogen in Enterobacter sp. MF84.








Similar content being viewed by others
Abbreviations
- ANOVA:
-
Analysis of variance
- CTAB:
-
Cetyltrimethylammonium Bromide
- LOF:
-
Lack of fit
- RSM:
-
Response surface methodology
References
Afzal I, Shinwari ZK, Iqrar I (2015) Selective isolation and characterization of agriculturally beneficial endophytic bacteria from wild hemp using canola. Pak J Bot 47:1999–2008
Ahmed I, Zia MA, Hussain MA, Akram Z, Naveed MT, Nowrouzi A (2016) Bioprocessing of citrus waste peel for induced pectinase production by Aspergillus niger; its purification and characterization. J Radiat Res Appl Sci 9:148–154
Anuradha K, Padma PN, Venkateshwar S, Reddy G (2016) Mango juice clarification with polygalacturonase produced by Aspergillus awamori MTCC 9166-optimization of conditions. Int Food Res J 23:147
Basu S, Ghosh A, Bera A, Saha MN, Chattopadhyay D, Chakrabarti K (2008) Thermodynamic characterization of a highly thermoactive extracellular pectate lyase from a new isolate Bacillus pumilus DKS1. Bioresour Technol 99:8088–8094
Beulah D, Sunitha E, Srilakshmi T (2015) Production, purification and assay of pectinase enzyme from Aspergillus niger. Helix 2:673–677
Bibi N, Ali S, Tabassum R (2016) Statistical optimization of pectinase biosynthesis from orange peel by Bacillus licheniformis using submerged fermentation. Waste Biomass Valori 7:467–481
Blanco P, Sieiro C, Villa TG (1999) Production of pectic enzymes in yeasts. FEMS Microbiol Lett 175:1–9
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brühlmann F, Kim KS, Zimmerman W, Fiechter A (1994) Pectinolytic enzymes from actinomycetes for the degumming of ramie bast fibers. Appl Environ Microbiol 60:2107–2112
Chandra Reddy MP, Saritha K (2016) Effects of the culture media optimization on pectinase production by Enterobacter sp. PSTB-1. 3 Biotech 6:207
Darah I, Nisha M, Lim S (2013) Enhancement of polygalacturonase production from Enterobacter aerogenes NBO2 by submerged fermentation. Adv Stud Biol 5:173–189
Handa S, Sharma N, Pathania S (2016) Multiple parameter optimization for maximization of pectinase production by Rhizopus sp. C4 under solid state fermentation. Fermentation 2:10
Hoondal G, Tiwari R, Tewari R, Dahiya N, Beg Q (2002) Microbial alkaline pectinases and their industrial applications: a review. Appl Microbiol Biotechnol 59:409–418
Huang LK, Mahoney R (1999) Purification and characterization of an endo-polygalacturonase from Verticillium albo-atrum. J Appl Microbiol 86:145–156
Jayani RS, Saxena S, Gupta R (2005) Microbial pectinolytic enzymes: a review. Process Biochem 40:2931–2944
Joshi M, Nerurkar M, Adivarekar R (2015) Characterization, kinetic, and thermodynamic studies of marine pectinase from Bacillus subtilis. Prep Biochem Biotechnol 45:205–220
Kashyap D, Vohra P, Chopra S, Tewari R (2001) Applications of pectinases in the commercial sector: a review. Bioresour Technol 77:215–227
Kaur SJ, Gupta VK (2017) Production of pectinolytic enzymes pectinase and pectin lyase by Bacillus subtilis SAV-21 in solid state fermentation. Ann Microbiol 67:333–342
Khataee AR, Zarei M, Asl SK (2010) Photocatalytic treatment of a dye solution using immobilized TiO2 nanoparticles combined with photoelectro-Fenton process: optimization of operational parameters. J Electroanal Chem 648:143–150
Khatri BP, Bhattarai T, Shrestha S, Maharjan J (2015) Alkaline thermostable pectinase enzyme from Aspergillus niger strain MCAS2 isolated from Manaslu Conservation Area, Gorkha, Nepal. Springerplus 4:488
Klug-Santner BG, Schnitzhofer W, Vrsanská M, Weber J, Agrawal PB, Nierstrasz VA, Guebitz GM (2006) Purification and characterization of a new bioscouring pectate lyase from Bacillus pumilus BK2. J Biotechnol 121:390–401
Kohli P, Gupta R (2015) Alkaline pectinases: a review. Biocatal Agric Biotechnol 4:279–285
Lara-Márquez A, Zavala-Páramo MG, López-Romero E, Camacho HC (2011) Biotechnological potential of pectinolytic complexes of fungi. Biotechnol Lett 33:859–868
Li P-J et al (2015) Optimizing production of pectinase from orange peel by Penicillium oxalicum PJ02 using response surface methodology. Waste Biomass Valoriz 6:13–22
Mahdinia E, Demirci A, Berenjian A (2019) Effects of medium components in a glycerol-based medium on vitamin K (menaquinone-7) production by Bacillus subtilis natto in biofilm reactors. Bioprocess Biosyst Eng 42:223–232
Maleki MH, Ghanbary MAT, Ranjbar G, Asgharzadeh A, Lotfi A (2017) Screening of some Zygomycetes strains for pectinase activity. J Microbiol Biotechnol Res 1:1–7
Martos MA, Zubreski ER, Garro OA, Hours RA (2013) Production of Pectinolytic enzymes by the yeast Wickerhanomyces anomalus isolated from citrus fruits peels. Biotechnol Res Int 2013: 435154
Mathew A, Eldo AN, Molly A (2008) Optimization of culture conditions for the production of thermostable polygalacturonase by Penicillium SPC-F 20. J Ind Microbiol Biotechnol 35:1001–1005
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Nawaz MA, Bhattarai RR (2015) Isolation and identification of Yersinia and Pseudomonas sp. from Australian milk and salad using 16s rDNA. In PeerJ PrePrints 3:e820v1
Padhi SK, Tripathy S, Mohanty S, Maiti NK (2017) Aerobic and heterotrophic nitrogen removal by Enterobacter cloacae CF-S27 with efficient utilization of hydroxylamine. Bioresour Technol 232:285–296
Patil SR, Dayanand A (2006) Optimization of process for the production of fungal pectinases from deseeded sunflower head in submerged and solid-state conditions. Bioresour Technol 97:2340–2344
Pedrolli DB, Monteiro AC, Gomes E, Carmona EC (2009) Pectin and pectinases: production, characterization and industrial application of microbial pectinolytic enzymes. Open Biotechnol J 3:9–18
Purnachandra Reddy M, Saritha K (2014) Studies on pectinase production by Enterobacter sp. using mango fruit processing industrial waste as whole and sole carbon source. Curr Trends Biotechnol Pharm 8:378–386
Rehman HU, Aman A, Nawaz MA, Qader SAU (2015) Characterization of pectin degrading polygalacturonase produced by Bacillus licheniformis KIBGE-IB21. Food Hydrocoll 43:819–824
Schaad NW, Jones JB, Chun W (2001) Laboratory guide for the identification of plant pathogenic bacteria. 3rd edn. American Phytopathological Society (APS Press). Saint Paul, Minnesota, USA.
Shah KP, Chandok KH, Rathore P, Sharma MV, Yadav M, Nayarisseri SA (2013) Screening, isolation and identification of polygalacturonase producing Bacillus tequilensis strain EMBS083 using 16S rRNA gene sequencing. Eur J Biol Sci 5:09–13
Sharma A, Husain I (2015) Optimization of medium components for extracellular glutaminase free asparaginase from Enterobacter cloacae. Int J Curr Microbiol App Sci 4:296–309
Sharma N, Rathore M, Sharma M (2013) Microbial pectinase: sources, characterization and applications. Rev Environ Sci Biotechnol 12:45–60
Singh S, Mandal SK (2012) Optimization of processing parameters for production of pectinolytic enzymes from fermented pineapple residue of mixed Aspergillus species. Jordan J Biol Sci 147:1–7
Sohail M, Latif Z (2016) Phylogenetic analysis of polygalacturonase-producing Bacillus and Pseudomonas isolated from plant waste material. Jundishapur J Microbiol 9:e28594
Thangaratham T, Manimegalai G (2014) Optimization and production of pectinase using agro waste by solid state and submerged fermentation. Int J Curr Microbiol App Sci 3:357–365
Uzuner S, Cekmecelioglu D (2015) Enhanced pectinase production by optimizing fermentation conditions of Bacillus subtilis growing on hazelnut shell hydrolyzate. J Mol Catal B Enzym 113:62–67
Venzon SS, Canteri MH, Granato D, Junior BD, Maciel GM, Stafussa AP, Haminiuk CW (2015) Physicochemical properties of modified citrus pectins extracted from orange pomace. J Food Sci Technol 52:4102–4112
Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey, FA, Schleifer, K-H, Whitman, WB (2011) Bergey's manual of systematic bacteriology: Volume 3: The Firmicutes. Springer Science & Business Media, Heidelberg, Germany.
Yadav S, Yadav P, Yadav D, Yadav K (2009) Pectin lyase: a review. Process Biochem 44:1–10
Yuan P, Meng K, Shi P, Luo H, Huang H, Tu T, Yang P, Yao B (2012) An alkaline-active and alkali-stable pectate lyase from Streptomyces sp. S27 with potential in textile industry. J Ind Microbiol Biotechnol 39:909–915
Zhou M, Wu J, Wang T, Gao L, Yin H, Lü X (2017) The purification and characterization of a novel alkali-stable pectate lyase produced by Bacillus subtilis PB1. World J Microbiol Biotechnol 33:190
Acknowledgments
The authors express their gratitude to the research council of Azarbaijan Shahid Madani University for the financial support during the course of this project.
Funding
This work was supported by the research council of Azarbaijan Shahid Madani University (Tabriz, Iran).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Research involving human participants and/or animals
Not applicable.
Informed consent
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Abdollahzadeh, R., Pazhang, M., Najavand, S. et al. Screening of pectinase-producing bacteria from farmlands and optimization of enzyme production from selected strain by RSM. Folia Microbiol 65, 705–719 (2020). https://doi.org/10.1007/s12223-020-00776-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-020-00776-7