Abstract
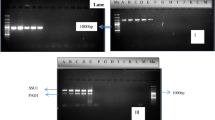
Yeast Candida utilis is considered to be a potentially advantageous expression system for production of recombinant proteins utilizable for industrial and pharmaceutical purposes. As the scientific literature is not consistent in the ploidy of this yeast, in this work, we focused on resolving the problem via several methods such as the copy number determination of maltase gene by multiplex PCR, measuring α-glucosidase activity, the characterization of maltase gene copy number in deletion mutants using qPCR and flow cytometry. In context with the published data and results obtained in this study about the copy number of the maltase gene on C. utilis genome, we attempted to hypothesise and made conclusion about the ploidy of C. utilis. The results of this work, besides the biotechnological aspect, contribute to the elementary knowledge of C. utilis. The exact information about the ploidy or more specifically about the copy number of appropriate gene is essential for expression cassette dosage determination integrated into the chromosome of the host. In this study, we come to the conclusion that the maltase gene is present in C. utilis genome in four alleles, and in combination with flow cytometry, published information and the published genome sequences, the observations support the theory about tetraploidy of C. utilis.







Similar content being viewed by others
References
Abad S, Kitz K, Hörmann A, Schreiner U, Hartner FS, Glieder A (2010) Real-time PCR-based determination of gene copy numbers in Pichia pastoris. Biotechnol J 5:413–420
Alamae T, Parn P, Viigand K, Karp H (2003) Regulation of the Hansenula polymorpha maltase gene promoter in H. polymorpha and Saccharomyces cerevisiae1. FEMS Yeast Res 4:165–173
Alves-Araujo C, Hernandez-Lopez MJ, Sousa MJ, Prieto JA, Randez-Gil F (2004) Cloning and characterization of the MAL11 gene encoding a high-affinity maltose transporter from Torulaspora delbrueckii. FEMS Yeast Res 4:467–476
Becker DM, Guarente L (1991) [12] High-efficiency transformation of yeast by electroporation. Methods Enzymol 194:182–187
Boňková H, Osadská M, Krahulec J, Lišková V, Stuchlík S, Turňa J (2014) Upstream regulatory regions controlling the expression of the Candida utilis maltase gene. J Biotechnol 189:136–142
Boze H, Moulin G, Galzy P (1992) Production of food and fodder yeasts. Crit Rev Biotechnol 12:65–86
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Buerth C, Heilmann CJ, Klis FM, de Koster CG, Ernst JF, Tielker D (2011) Growth-dependent secretome of Candida utilis. Microbiology 157:2493–2503. https://doi.org/10.1099/mic.0.049320-0
Chou WY, Marky LA, Zaunczkowski D, Breslauer KJ (1987) The thermodynamics of drug-DNA interactions: ethidium bromide and propidium iodine. J Biomol Struct Dyn 5(2):345–359
Cuomo CA, Fanning S, Gujja S, Zeng Q, Naglik JR, Filler SG, Mitchell AP (2019) Genome sequence for Candida albicans clinical oral isolate 529L. Microbiol Resour Announc 8(25). https://doi.org/10.1128/MRA.00554-19
Darzynkiewicz Z, Huang X, Zhao H (2017) Analysis of cellular DNA content by flow cytometry. Curr Protoc Immunol 119:5.7.1–5.7.20
Geber A, Williamson PR, Rex JH, Sweeney EC, Bennett JE (1992) Cloning and characterization of a Candida albicans maltase gene involved in sucrose utilization. J Bacteriol 174:6992–6996
Goldstein AL, McCusker JH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553
Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24:2519–2524
Ikushima S, Fujii T, Kobayashi O (2009) Efficient gene disruption in the high-ploidy yeast Candida utilis using the Cre-loxP system. Biosci Biotechnol Biochem 73:879–884. https://doi.org/10.1271/bbb.80799
Kondo K, Saito T, Kajiwara S, Takagi M, Misawa N (1995) A transformation system for the yeast Candida utilis: use of a modified endogenous ribosomal protein gene as a drug-resistant marker and ribosomal DNA as an integration target for vector DNA. J Bacteriol 177:7171–7177
Kondo K, Miura Y, Sone H, Kobayashi K, Iijima H (1997) High-level expression of a sweet protein, monellin, in the food yeast Candida utilis. Nat Biotechnol 15:453–457. https://doi.org/10.1038/nbt0597-453
Krahulec J, Boňková H, Lišková V, Osadská M, Stuchlík S, Turňa J (2014) Deletion of the maltase gene regulatory regions in yeast candida utilis. FEBS J 281:680
Kunigo M, Buerth C, Tielker D, Ernst JF (2013) Heterologous protein secretion by Candida utilis. Appl Microbiol Biotechnol 97:7357–7368
Kurtzman CP, Johnson CJ, Smiley MJ (1979) Determination of conspecificity of Candida utilis and Hansenula jadinii through DNA reassociation. Mycologia 11:844–847
Madinger CL et al (2009) The effect of carbon source on the secretome of Kluyveromyces lactis. Proteomics 9:4744–4754. https://doi.org/10.1002/pmic.200800915
Miura Y, Kondo K, Saito T, Shimada H, Fraser PD, Misawa N (1998) Production of the carotenoids lycopene, beta-carotene, and astaxanthin in the food yeast Candida utilis. Appl Environ Microbiol 64:1226–1229
Miura Y, Kettoku M, Kato M, Kobayashi K, Kondo K (1999) High level production of thermostable alpha-amylase from Sulfolobus solfataricus in high-cell density culture of the food yeast Candida utilis. J Mol Microbiol Biotechnol 1:129–134
Needleman R (1991) Control of maltase synthesis in yeast. Mol Microbiol 5:2079–2084
Porro D, Sauer M, Branduardi P, Mattanovich D (2005) Recombinant protein production in yeasts. Mol Biotechnol 31:245–259
Rodríguez L, Chávez FP, Basabe L, Rivero T, Delgado JM (1998) Development of an integrative DNA transformation system for the yeast Candida utilis. FEMS Microbiol Lett 165:335–340
Rolim MF, de Araujo PS, Panek AD, Paschoalin VM, Silva JT (2003) Shared control of maltose and trehalose utilization in Candida utilis. Braz J Med Biol Res 36:829–837
Rose MD, Winston FM, Heiter P (1990) Methods in yeast genetics: a laboratory course manual
Rupp O, Brinkrolf K, Buerth C, Kunigo M, Schneider J, Jaenicke S, Goesmann A, Puhler A, Jaeger KE, Ernst JF (2015) The structure of the Cyberlindnera jadinii genome and its relation to Candida utilis analyzed by the occurrence of single nucleotide polymorphisms. J Biotechnol 211:20–30. https://doi.org/10.1016/j.jbiotec.2015.06.423
Sambrook J, Fritsch E, Maniatis T (1989) In: Sambrook J, Fritsch EF, Maniatis T (eds) Molecular cloning : a laboratory manual. Cold Spring Harbor laboratory press, New York
Sartory A, Sartory R, Weill J, Meyer J (1932) Un cas de blastomycose invétérée transmissible au cobaye, due à un Saccharomyces pathogène (Saccharomyces jadini n. sp.). Comptes Rendues des Séances Hebdomadaires de l'Académie des Sciences Paris 194:1688–1690
Shimada H, Kondo K, Fraser PD, Miura Y, Saito T, Misawa N (1998) Increased carotenoid production by the food yeast Candida utilis through metabolic engineering of the isoprenoid pathway. Appl Environ Microbiol 64:2676–2680
Shimada H, Obayashi T, Takahashi N, Matsui M, Sakamoto A (2010) Normalization using ploidy and genomic DNA copy number allows absolute quantification of transcripts, proteins and metabolites in cells. Plant Methods 6:1
Tomita Y, Ikeo K, Tamakawa H, Gojobori T, Ikushima S (2012) Genome and transcriptome analysis of the food-yeast Candida utilis. PLoS One 7:e37226. https://doi.org/10.1371/journal.pone.0037226
Wei W, Hong-Lan Y, HuiFang B, Daoyuan Z, Wood AJ (2010) The effective expression of xylanase gene in Candida utilis by 18S rDNA targeted homologous recombination in pGLR9K. Mol Biol Rep 37:2615–2620
Wilson RB, Davis D, Mitchell AP (1999) Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181:1868–1874
Yamada Y, Matsuda M, Mikata K (1995) The phylogenetic relationships of Pichia jadinii, formerly classified in the genus Hansenula, and related species based on the partial sequences of 18S and 26S ribosomal RNAs (Saccharomycetaceae). Biosci Biotechnol Biochem 59:518–520
Contributors
The author Ján Krahulec declares that he participated in the article preparation. The author Veronika Lišková declares that she materially participated in the research and also participated in the article preparation. The author Hana Boňková declares that she materially participated in the research and also participated in the article preparation. The author Aneta Lichvariková declares that she materially participated in the research. Martin Šafranek declares that he materially participated in the research. The author Ján Turňa declares that he participated in the article preparation.
Funding
This work is a result of project implementation “Production of biologically active agents based on recombinant proteins” (ITMS 26240220048) supported by the Research and Development Operational Program, funded by the ERDF, and also supported by the grant of Slovak research and development agency APVV-17-0149.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by: Agnieszka Szalewska-Palasz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Krahulec, J., Lišková, V., Boňková, H. et al. The ploidy determination of the biotechnologically important yeast Candida utilis. J Appl Genetics 61, 275–286 (2020). https://doi.org/10.1007/s13353-020-00544-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-020-00544-w