Abstract
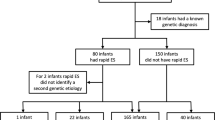
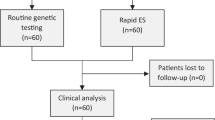
Genome sequencing is used to make genetic diagnoses in critically ill infants with rapid turnaround time (TAT). Herein, to delineate the value of a genetic diagnosis, we provide the results from 130 pediatric patients in a large, comprehensive children’s hospital in China. This study was performed using an optimized trio genome sequencing (OTGS) test. The sequencing depth for patients was 40–50 × and for their parents, it was 8–10 × . Patients from the pediatric or neonatal intensive care unit (PICU/NICU) with complicated clinical features were enrolled between June 2018 and December 2018, each with a phenotype suggesting an underlying genetic disorder. OTGS testing identified pathogenic variants in 62 of 130 individuals, resulting in a diagnosis rate of 47.7%. The TAT varied from 72 to 120 h, with an average of 94 h and a median of 90 h. Of the 62 infants with diagnoses, 48 (77.4%) had pathogenic single-nucleotide variants (SNVs), 12 (19.4%) had pathogenic copy number variations (CNVs) or structure variants (SVs), and 2 (3.2%) had small deletions in one allele plus pathogenic variants in another allele of autosomal recessive genes. Therapeutic strategies for 48.4% (30/62) of the diagnosed patients were modified and included transplantation, dietary recommendations, or change of drugs, which avoided morbidity and improved prognosis. This study provided high-capacity OTGS testing in detecting SNVs and chromosomal abnormalities with fast response, higher diagnostic yield, and lower cost. OTGS demonstrates the potential to be the first-tier of genetic testing used in critically ill infants in developing countries.

Similar content being viewed by others
References
Alfares A, Aloraini T, Subaie LA, Alissa A, Qudsi AA, Alahmad A, Mutairi FA, Alswaid A, Alothaim A, Eyaid W, Albalwi M, Alturki S, Alfadhel M (2018) Whole-genome sequencing offers additional but limited clinical utility compared with reanalysis of whole-exome sequencing. Genet Med 20:1328–1333. https://doi.org/10.1038/gim.2018.41
Anderson S, Koniaris S, Xin B, Brooks SS (2017) Congenital glucose-galactose malabsorption: a case report. J Pediatr Health Care 31:506–510. https://doi.org/10.1016/j.pedhc.2017.01.005
Farnaes L, Hildreth A, Sweeney NM, Clark MM, Chowdhury S, Nahas S, Cakici JA, Benson W, Kaplan RH, Kronick R, Bainbridge MN, Friedman J, Gold JJ, Ding Y, Veeraraghavan N, Dimmock D, Kingsmore SF (2018) Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom Med 3:10. https://doi.org/10.1038/s41525-018-0049-4
French CE, Delon I, Dolling H, Sanchis-Juan A, Shamardina O, Megy K, Abbs S, Austin T, Bowdin S, Branco RG, Firth H, Disease NB-R, Next Generation Children P, Rowitch DH, Raymond FL (2019) Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med 45:627–636. https://doi.org/10.1007/s00134-019-05552-x
Gross AM, Ajay SS, Rajan V, Brown C, Bluske K, Burns NJ, Chawla A, Coffey AJ, Malhotra A, Scocchia A, Thorpe E, Dzidic N, Hovanes K, Sahoo T, Dolzhenko E, Lajoie B, Khouzam A, Chowdhury S, Belmont J, Roller E, Ivakhno S, Tanner S, McEachern J, Hambuch T, Eberle M, Hagelstrom RT, Bentley DR, Perry DL, Taft RJ (2019) Copy-number variants in clinical genome sequencing: deployment and interpretation for rare and undiagnosed disease. Genet Med 21:1121–1130. https://doi.org/10.1038/s41436-018-0295-y
Hu X, Li N, Xu Y, Li G, Yu T, Yao RE, Fu L, Wang J, Yin L, Yin Y, Wang Y, Jin X, Wang X, Wang J, Shen Y (2018) Proband-only medical exome sequencing as a cost-effective first-tier genetic diagnostic test for patients without prior molecular tests and clinical diagnosis in a developing country: the China experience. Genet Med 20:1045–1053. https://doi.org/10.1038/gim.2017.195
Li Z, Zhang F, Wang Y, Qiu Y, Wu Y, Lu Y, Yang L, Qu WJ, Wang H, Zhou W, Tian W (2019) PhenoPro: a novel toolkit for assisting in the diagnosis of Mendelian disease. Bioinformatics. https://doi.org/10.1093/bioinformatics/btz100
Lionel AC, Costain G, Monfared N, Walker S, Reuter MS, Hosseini SM, Thiruvahindrapuram B, Merico D, Jobling R, Nalpathamkalam T, Pellecchia G, Sung WWL, Wang Z, Bikangaga P, Boelman C, Carter MT, Cordeiro D, Cytrynbaum C, Dell SD, Dhir P, Dowling JJ, Heon E, Hewson S, Hiraki L, Inbar-Feigenberg M, Klatt R, Kronick J, Laxer RM, Licht C, MacDonald H, Mercimek-Andrews S, Mendoza-Londono R, Piscione T, Schneider R, Schulze A, Silverman E, Siriwardena K, Snead OC, Sondheimer N, Sutherland J, Vincent A, Wasserman JD, Weksberg R, Shuman C, Carew C, Szego MJ, Hayeems RZ, Basran R, Stavropoulos DJ, Ray PN, Bowdin S, Meyn MS, Cohn RD, Scherer SW, Marshall CR (2018) Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med 20:435–443. https://doi.org/10.1038/gim.2017.119
Longoni M, High FA, Qi H, Joy MP, Hila R, Coletti CM, Wynn J, Loscertales M, Shan L, Bult CJ, Wilson JM, Shen Y, Chung WK, Donahoe PK (2017) Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Hum Genet 136:679–691. https://doi.org/10.1007/s00439-017-1774-y
Matthijs G, Souche E, Alders M, Corveleyn A, Eck S, Feenstra I, Race V, Sistermans E, Sturm M, Weiss M, Yntema H, Bakker E, Scheffer H, Bauer P, EuroGentest, European Society of Human G (2016) Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet 24:2–5. https://doi.org/10.1038/ejhg.2015.226
Meienberg J, Bruggmann R, Oexle K, Matyas G (2016) Clinical sequencing: is WGS the better WES? Hum Genet 135:359–362. https://doi.org/10.1007/s00439-015-1631-9
Meng L, Pammi M, Saronwala A, Magoulas P, Ghazi AR, Vetrini F, Zhang J, He W, Dharmadhikari AV, Qu C, Ward P, Braxton A, Narayanan S, Ge X, Tokita MJ, Santiago-Sim T, Dai H, Chiang T, Smith H, Azamian MS, Robak L, Bostwick BL, Schaaf CP, Potocki L, Scaglia F, Bacino CA, Hanchard NA, Wangler MF, Scott D, Brown C, Hu J, Belmont JW, Burrage LC, Graham BH, Sutton VR, Craigen WJ, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Muzny DM, Miller MJ, Wang X, Leduc MS, Xiao R, Liu P, Shaw C, Walkiewicz M, Bi W, Xia F, Lee B, Eng CM, Yang Y, Lalani SR (2017) Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr 171:e173438. https://doi.org/10.1001/jamapediatrics.2017.3438
Mestek-Boukhibar L, Clement E, Jones WD, Drury S, Ocaka L, Gagunashvili A, Le Quesne Stabej P, Bacchelli C, Jani N, Rahman S, Jenkins L, Hurst JA, Bitner-Glindzicz M, Peters M, Beales PL, Williams HJ (2018) Rapid Paediatric Sequencing (RaPS): comprehensive real-life workflow for rapid diagnosis of critically ill children. J Med Genet 55:721–728. https://doi.org/10.1136/jmedgenet-2018-105396
Miller NA, Farrow EG, Gibson M, Willig LK, Twist G, Yoo B, Marrs T, Corder S, Krivohlavek L, Walter A, Petrikin JE, Saunders CJ, Thiffault I, Soden SE, Smith LD, Dinwiddie DL, Herd S, Cakici JA, Catreux S, Ruehle M, Kingsmore SF (2015) A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med 7:100. https://doi.org/10.1186/s13073-015-0221-8
Petersen BS, Fredrich B, Hoeppner MP, Ellinghaus D, Franke A (2017) Opportunities and challenges of whole-genome and -exome sequencing. BMC Genet 18:14. https://doi.org/10.1186/s12863-017-0479-5
Petrikin JE, Cakici JA, Clark MM, Willig LK, Sweeney NM, Farrow EG, Saunders CJ, Thiffault I, Miller NA, Zellmer L, Herd SM, Holmes AM, Batalov S, Veeraraghavan N, Smith LD, Dimmock DP, Leeder JS, Kingsmore SF (2018) The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genom Med 3:6. https://doi.org/10.1038/s41525-018-0045-8
Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, Humphray S, Saffrey P, Kingsbury Z, Weir JC, Betley J, Grocock RJ, Margulies EH, Farrow EG, Artman M, Safina NP, Petrikin JE, Hall KP, Kingsmore SF (2012) Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med 4:154ra135. https://doi.org/10.1126/scitranslmed.3004041
Schwarze K, Buchanan J, Taylor JC, Wordsworth S (2018) Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med 20:1122–1130. https://doi.org/10.1038/gim.2017.247
Smith LD, Willig LK, Kingsmore SF (2015) Whole-exome sequencing and whole-genome sequencing in critically ill neonates suspected to have single-gene disorders. Cold Spring Harb Perspect Med 6:a023168. https://doi.org/10.1101/cshperspect.a023168
Smith HS, Swint JM, Lalani SR, Yamal JM, de Oliveira Otto MC, Castellanos S, Taylor A, Lee BH, Russell HV (2019) Clinical application of genome and exome sequencing as a diagnostic tool for pediatric patients: a scoping review of the literature. Genet Med 21:3–16. https://doi.org/10.1038/s41436-018-0024-6
Stranneheim H, Wedell A (2016) Exome and genome sequencing: a revolution for the discovery and diagnosis of monogenic disorders. J Intern Med 279:3–15. https://doi.org/10.1111/joim.12399
Stray-Pedersen A, Sorte HS, Samarakoon P, Gambin T, Chinn IK, Coban Akdemir ZH, Erichsen HC, Forbes LR, Gu S, Yuan B, Jhangiani SN, Muzny DM, Rodningen OK, Sheng Y, Nicholas SK, Noroski LM, Seeborg FO, Davis CM, Canter DL, Mace EM, Vece TJ, Allen CE, Abhyankar HA, Boone PM, Beck CR, Wiszniewski W, Fevang B, Aukrust P, Tjonnfjord GE, Gedde-Dahl T, Hjorth-Hansen H, Dybedal I, Nordoy I, Jorgensen SF, Abrahamsen TG, Overland T, Bechensteen AG, Skogen V, Osnes LTN, Kulseth MA, Prescott TE, Rustad CF, Heimdal KR, Belmont JW, Rider NL, Chinen J, Cao TN, Smith EA, Caldirola MS, Bezrodnik L, Lugo Reyes SO, Espinosa Rosales FJ, Guerrero-Cursaru ND, Pedroza LA, Poli CM, Franco JL, Trujillo Vargas CM, Aldave Becerra JC, Wright N, Issekutz TB, Issekutz AC, Abbott J, Caldwell JW, Bayer DK, Chan AY, Aiuti A, Cancrini C, Holmberg E, West C, Burstedt M, Karaca E, Yesil G, Artac H, Bayram Y, Atik MM, Eldomery MK, Ehlayel MS, Jolles S, Flato B, Bertuch AA, Hanson IC, Zhang VW, Wong LJ, Hu J, Walkiewicz M, Yang Y, Eng CM, Boerwinkle E, Gibbs RA, Shearer WT, Lyle R, Orange JS, Lupski JR (2017) Primary immunodeficiency diseases: genomic approaches delineate heterogeneous Mendelian disorders. J Allergy Clin Immunol 139:232–245. https://doi.org/10.1016/j.jaci.2016.05.042
Tarailo-Graovac M, Shyr C, Ross CJ, Horvath GA, Salvarinova R, Ye XC, Zhang LH, Bhavsar AP, Lee JJ, Drogemoller BI, Abdelsayed M, Alfadhel M, Armstrong L, Baumgartner MR, Burda P, Connolly MB, Cameron J, Demos M, Dewan T, Dionne J, Evans AM, Friedman JM, Garber I, Lewis S, Ling J, Mandal R, Mattman A, McKinnon M, Michoulas A, Metzger D, Ogunbayo OA, Rakic B, Rozmus J, Ruben P, Sayson B, Santra S, Schultz KR, Selby K, Shekel P, Sirrs S, Skrypnyk C, Superti-Furga A, Turvey SE, Van Allen MI, Wishart D, Wu J, Wu J, Zafeiriou D, Kluijtmans L, Wevers RA, Eydoux P, Lehman AM, Vallance H, Stockler-Ipsiroglu S, Sinclair G, Wasserman WW, van Karnebeek CD (2016) Exome sequencing and the management of neurometabolic disorders. N Engl J Med 374:2246–2255. https://doi.org/10.1056/NEJMoa1515792
van Diemen CC, Kerstjens-Frederikse WS, Bergman KA, de Koning TJ, Sikkema-Raddatz B, van der Velde JK, Abbott KM, Herkert JC, Lohner K, Rump P, Meems-Veldhuis MT, Neerincx PBT, Jongbloed JDH, van Ravenswaaij-Arts CM, Swertz MA, Sinke RJ, van Langen IM, Wijmenga C (2017) Rapid targeted genomics in critically ill newborns. Pediatrics. https://doi.org/10.1542/peds.2016-2854
Willig LK, Petrikin JE, Smith LD, Saunders CJ, Thiffault I, Miller NA, Soden SE, Cakici JA, Herd SM, Twist G, Noll A, Creed M, Alba PM, Carpenter SL, Clements MA, Fischer RT, Hays JA, Kilbride H, McDonough RJ, Rosterman JL, Tsai SL, Zellmer L, Farrow EG, Kingsmore SF (2015) Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med 3:377–387. https://doi.org/10.1016/S2213-2600(15)00139-3
Yang L, Kong Y, Dong X, Hu L, Lin Y, Chen X, Ni Q, Lu Y, Wu B, Wang H, Lu QR, Zhou W (2018) Clinical and genetic spectrum of a large cohort of children with epilepsy in China. Genet Med. https://doi.org/10.1038/s41436-018-0091-8
Acknowledgements
We thank the clinicians for taking care of the patients and our genetics laboratory teams who contributed to this study. We also thank Dr. Fan Xia of Baylor Medical School, who gave us suggestions for the method of OTGS and guidance for the manuscript. The sequencing reagents were funded by the Ministry of Science and Technology National Key Research and Development Program (2018YFC0116903), Shen Kang Hospital Development Center Clinical Science and Technology Innovation Project of Shanghai (SHDC12017110), and Shanghai health and family planning commission (201440628). The sequencing platform was supported by the project of Shanghai Key Laboratory of Birth Defects (13DZ2260600). The corresponding author had full access to all the data in this study and had final responsibility for the decision to submit for publication.
Author information
Authors and Affiliations
Contributions
WHZ conceptualized and designed the study, coordinated the study overall, and revised the manuscript; HJW co-designed the study, drafted the initial manuscript, and revised the manuscript; BBW developed the laboratory, interpreted data, and critically reviewed the manuscript; LY enrolled patients and was involved in clinical discussions with clinicians. GPL and GQQ treated patients in the ICU and discussed the genetic results with multidisciplinary teams. YLL and XRD developed the algorithm and data analysis pipeline and revised the manuscript; YYQ performed experiments and collected and interpreted data; QN helped to collect and summarize data; PZ performed laboratory work and analyzed and interpreted data. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, H., Lu, Y., Dong, X. et al. Optimized trio genome sequencing (OTGS) as a first-tier genetic test in critically ill infants: practice in China. Hum Genet 139, 473–482 (2020). https://doi.org/10.1007/s00439-019-02103-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-019-02103-8