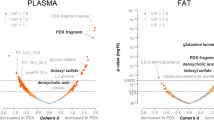
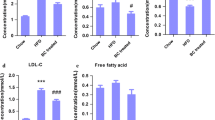
Abstract
This study aimed to evaluate the effect of probiotic administration on obese and ageing models. Sprague Dawley rats were subjected to high-fat diet (HFD) and injected with D-galactose to induce premature ageing. Upon 12 weeks of treatment, the faecal samples were collected and subjected to gas chromatography–mass spectrophotometry (GC-MS) analysis for metabolite detection. The sparse partial least squares discriminant analysis (sPLS-DA) showed a distinct clustering pattern of metabolite profile in the aged and obese rats administered with probiotics Lactobacillus plantarum DR7 and L. reuteri 8513d, particularly with a significantly higher concentration of allantoin. Molecular docking simulation showed that allantoin promoted the phosphorylation (activation) of adenosine monophosphate-activated kinase (AMPK) by lowering the substrate free energy of binding (FEB) and induced the formation of an additional hydrogen bond between Val184 and the substrate AMP. Allantoin also suppressed cholesterol biosynthesis by either inducing enzyme inhibition, occupying or blocking the putative binding site to result in non-spontaneous substrate binding, as in the cases of 3-hydroxy-methylglutaryl-coA reductase (HMGCR), mevalonate kinase (MVK) and lanosterol demethylase (LDM) where positive FEBs were reported. These results demonstrated the potential of allantoin to alleviate age-related hypercholesterolaemia by upregulating AMPK and downregulating cholesterol biosynthesis via the mevalonate pathway and Bloch pathway.








Similar content being viewed by others
References
Houston, D. K., Nicklas, B. J., & Zizza, C. A. (1886-1895). Weighty concerns: the growing prevalence of obesity among older adults. Journal of the American Dietetic Association, 2009, 109.
Feingold, K.R.; Grunfeld, C. (2018). Obesity and dyslipidemia. In Endotext [Internet], MDText. com, Inc.
Nelson, R. H. (2013). Hyperlipidemia as a risk factor for cardiovascular disease. Primary Care; Clinics in Office Practice, 40(1), 195–211.
Mazein, A., Watterson, S., Hsieh, W.-Y., Griffiths, W. J., & Ghazal, P. (2013). A comprehensive machine-readable view of the mammalian cholesterol biosynthesis pathway. Biochemical Pharmacology, 86(1), 56–66.
Mitsche, M. A., McDonald, J. G., Hobbs, H. H., & Cohen, J. C. (2015). Flux analysis of cholesterol biosynthesis in vivo reveals multiple tissue and cell-type specific pathways. Elife, 4, 56–66.
Hardie, D. G. (2011). AMP-activated protein kinase—an energy sensor that regulates all aspects of cell function. Genes & Development, 25, 1895–1908.
Mihaylova, M. M., & Shaw, R. J. (2011). The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature Cell Biology, 13, 1016.
Salminen, A., & Kaarniranta, K. (2012). AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Research Reviews, 11(2), 230–241.
Steinberg, G. R., & Kemp, B. E. (2009). AMPK in health and disease. Physiological Reviews, 89(3), 1025–1078.
Kerry, R. G., Patra, J. K., Gouda, S., Park, Y., Shin, H.-S., & Das, G. (2018). Benefaction of probiotics for human health: a review. Journal of Food and Drug Analysis, 26, 927–939.
Sánchez, B., Delgado, S., Blanco-Míguez, A., Lourenço, A., Gueimonde, M., & Margolles, A. (2017). Probiotics, gut microbiota, and their influence on host health and disease. Molecular Nutrition & Food Research, 61, 1600240.
Vaiserman, A. M., Koliada, A. K., & Marotta, F. (2017). Gut microbiota: a player in aging and a target for anti-aging intervention. Ageing Research Reviews, 35, 36–45.
Lew, L. C., Hor, Y. Y., Jaafar, M. H., Khoo, B. Y., Sasidharan, S., Choi, S. B., Ong, K. L., Kato, T., Nakanishi, Y., Ohno, H., & Liong, M. T. (2019). Effects of potential probiotic strains on the fecal microbiota and metabolites of d-galactose-induced aging rats fed with high-fat diet. Probiotics and antimicrobial proteins, 1–18.
Xia, J., Sinelnikov, I. V., Han, B., & Wishart, D. S. (2015). MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Research, 43, W251–W257.
Kanehisa, M., & Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research, 28(1), 27–30.
Bernstein, F. C., Koetzle, T. F., Williams, G. J., Meyer Jr., E. F., Brice, M. D., Rodgers, J. R., Kennard, O., Shimanouchi, T., & Tasumi, M. (1977). The Protein Data Bank: a computer-based archival file for macromolecular structures. Journal of Molecular Biology, 112(3), 535–542.
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791.
Salentin, S., Schreiber, S., Haupt, V. J., Adasme, M. F., & Schroeder, M. (2015). PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Research, 43(W1), W443–W447.
Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: visual molecular dynamics. Journal of Molecular Graphics, 14, 33–38.
Laskowski, R. A., & Swindells, M. B. (2011). LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling, 51(10), 2778–2786.
De Preter, V., Ghebretinsae, A. H., Abrahantes, J. C., Windey, K., Rutgeerts, P., & Verbeke, K. (2011). Impact of the synbiotic combination of Lactobacillus casei Shirota and oligofructose-enriched inulin on the fecal volatile metabolite profile in healthy subjects. Molecular Nutrition & Food Research, 55, 714–722.
Nealon, N. J., Yuan, L., Yang, X., & Ryan, E. P. (2017). Rice bran and probiotics alter the porcine large intestine and serum metabolomes for protection against human rotavirus diarrhea. Frontiers in Microbiology, 8, 653.
Grandiosa, R., Mérien, F., Young, T., Van Nguyen, T., Gutierrez, N., Kitundu, E., & Alfaro, A. C. (2018). Multi-strain probiotics enhance immune responsiveness and alters metabolic profiles in the New Zealand black-footed abalone (Haliotis iris). Fish & Shellfish Immunology, 82, 330–338.
Nguyen, T. L., Chun, W.-K., Kim, A., Kim, N., Roh, H. J., Lee, Y., Yi, M., Park, C.-I., & Kim, D.-H. (2018). Dietary probiotic effect of Lactococcus lactis WFLU12 on low-molecular-weight metabolites and growth of olive flounder (Paralichythys olivaceus). Frontiers in Microbiology, 2059, 9.
Wang, W., Li, Z., Gan, L., Fan, H., & Guo, Y. (2018). Dietary supplemental Kluyveromyces marxianus alters the serum metabolite profile in broiler chickens. Food & Function, 9, 3776–3787.
Hor, Y.-Y., Lew, L.-C., Jaafar, M. H., Lau, A. S.-Y., Ong, J.-S., Kato, T., Nakanishi, Y., Azzam, G., Azlan, A., & Ohno, H. (2019). Lactobacillus sp. improved microbiota and metabolite profiles of aging rats. Pharmacological Research, 104312.
Vernocchi, P., Del Chierico, F., & Putignani, L. (2016). Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Frontiers in Microbiology, 7, 1144.
Thornfeldt, C. (2005). Cosmeceuticals containing herbs: fact, fiction, and future. Dermatologic Surgery, 31, 873–881.
Maiuolo, J., Oppedisano, F., Gratteri, S., Muscoli, C., & Mollace, V. (2016). Regulation of uric acid metabolism and excretion. International Journal of Cardiology, 213, 8–14.
El Mubarak, M. A. S., Lamari, F. N., & Kontoyannis, C. (2013). Simultaneous determination of allantoin and glycolic acid in snail mucus and cosmetic creams with high performance liquid chromatography and ultraviolet detection. Journal of Chromatography A, 1322, 49–53.
Araújo, L. U., Grabe-Guimarães, A., Mosqueira, V. C. F., Carneiro, C. M., & Silva-Barcellos, N. M. (2010). Profile of wound healing process induced by allantoin. Acta Cirúrgica Brasileira, 25, 460–461.
Go, H.-K., Rahman, M., Kim, G.-B., Na, C.-S., Song, C.-H., Kim, J.-S., Kim, S.-J., & Kang, H.-S. (2015). Antidiabetic effects of yam (Dioscorea batatas) and its active constituent, allantoin, in a rat model of streptozotocin-induced diabetes. Nutrients, 7, 8532–8544.
Lin, K.-C., Yeh, L.-R., Chen, L.-J., Wen, Y.-J., Cheng, K.-C., & Cheng, J.-T. (2012). Plasma glucose-lowering action of allantoin is induced by activation of imidazoline I-2 receptors in streptozotocin-induced diabetic rats. Hormone and Metabolic Research, 44(1), 41–46.
Niu, C.-S., Chen, W., Wu, H.-T., Cheng, K.-C., Wen, Y.-J., Lin, K.-C., & Cheng, J.-T. (2010). Decrease of plasma glucose by allantoin, an active principle of yam (Dioscorea spp.), in streptozotocin-induced diabetic rats. Journal of Agricultural and Food Chemistry, 58(22), 12031–12035.
Kim, H.-L., Jeon, Y.-D., Park, J., Rim, H.-K., Jeong, M.-Y., Lim, H., Ko, S.-G., Jang, H.-J., Lee, B.-C., & Lee, K.-T. (2013). Corni fructus containing formulation attenuates weight gain in mice with diet-induced obesity and regulates adipogenesis through AMPK. Evidence-based Complementary and Alternative Medicine, 2013.
Ma, J., Kang, S., Meng, X., Kang, A., Park, J., Park, Y.-K., & Jung, H. (2018). Effects of rhizome extract of Dioscorea batatas and its active compound, allantoin, on the regulation of myoblast differentiation and mitochondrial biogenesis in C2C12 myotubes. Molecules, 2023, 23.
Kahraman, A., Morris, R. J., Laskowski, R. A., Favia, A. D., & Thornton, J. M. (2010). On the diversity of physicochemical environments experienced by identical ligands in binding pockets of unrelated proteins. Proteins: Structure, Function, and Bioinformatics, 78, 1120–1136.
Istvan, E. S., Palnitkar, M., Buchanan, S. K., & Deisenhofer, J. (2000). Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. The EMBO Journal, 19, 819–830.
Liong, M.-T., Lee, B.-H., Choi, S.-B., Lew, L.-C., Lau, A.-S.-Y., & Daliri, B. (2015). Cholesterol-lowering effects of probiotics and prebiotics. Probiotics and Prebiotics, 429–446.
Friesen, J. A., & Rodwell, V. W. (2004). The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biology, 5, 248.
Thompson, P. D., Panza, G., Zaleski, A., & Taylor, B. (2016). Statin-associated side effects. Journal of the American College of Cardiology, 67, 2395–2410.
Fu, Z., Wang, M., Potter, D., Miziorko, H. M., & Kim, J.-J. P. (2002). The structure of a binary complex between a mammalian mevalonate kinase and ATP: insights into the reaction mechanism and human inherited disease. Journal of Biological Chemistry, 277(20), 18134–18142.
Dougherty, D. A. (2007). Cation-π interactions involving aromatic amino acids. The Journal of Nutrition, 137, 1504S–1508S.
Tu, T., Li, Y., Su, X., Meng, K., Ma, R., Wang, Y., Yao, B., Lin, Z., & Luo, H. (2016). Probing the role of cation-π interaction in the thermotolerance and catalytic performance of endo-polygalacturonases. Scientific Reports, 6, 38413.
Strushkevich, N., Usanov, S. A., & Park, H.-W. (2010). Structural basis of human CYP51 inhibition by antifungal azoles. Journal of Molecular Biology, 397(4), 1067–1078.
Acknowledgments
P.-G.Y. would like to thank M.-T.L. and S.-B.C. for providing guidance and support throughout the project.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yap, PG., Choi, SB. & Liong, MT. Allantoin, a Potential Metabolite That Promotes AMPK Phosphorylation and Suppresses Cholesterol Biosynthesis Via the Mevalonate Pathway and Bloch Pathway. Appl Biochem Biotechnol 191, 226–244 (2020). https://doi.org/10.1007/s12010-020-03265-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03265-2