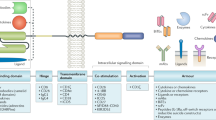
Abstract
The expression of synthetic receptors in primary T cells enables the programming of user-defined responses when designing T-cell therapies. Chimeric antigen receptors (CARs) are synthetic receptors that have demonstrated efficacy in cancer therapy by targeting immobilized antigens on the surface of malignant cells. Recently, we showed they can also rewire T-cell responses to soluble ligands. In contrast to other synthetic receptors, CARs are not only readily engineered by rational design, but also clinically translatable, with robust function in primary human T cells. This protocol discusses design principles for CARs responsive to soluble ligands and delineates steps for producing T cells expressing synthetic receptors. Functional assays for quantifying the ability of CAR T cells to sense and respond to soluble ligands are also presented. This protocol provides a framework for proficient immune-cell researchers to test novel T-cell therapies targeting soluble ligands in <2 weeks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data analyzed to generate what is shown in this manuscript are compiled in the Supplementary Spreadsheet file. Individual examples of gating strategy are shown when appropriate. Data from Fig. 4 and Supplementary Fig. 3 have been used in a prior publication6.
References
Maude, S. L. et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371, 1507–1517 (2014).
Maude, S. L. et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018).
Porter, D. L. et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med. 7, 303ra139 (2015).
Brentjens, R. J. et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood 118, 4817–4828 (2011).
Kochenderfer, J. N. et al. Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J. Clin. Oncol. 33, 540–549 (2015).
Chang, Z. L. et al. Rewiring T-cell responses to soluble factors with chimeric antigen receptors. Nat. Chem. Biol. 14, 317–324 (2018).
Hou, A. J., Chang, Z. L., Lorenzini, M. H., Zah, E. & Chen, Y. Y. TGF‐β–responsive CAR‐T cells promote anti‐tumor immune function. Bioeng. Transl. Med 3, 75–86 (2018).
Chen, L. C. & Chen, Y. Y. Outsmarting and outmuscling cancer cells with synthetic and systems immunology. Curr. Opin. Biotechnol. 60, 111–118 (2019).
Leen, A. M. et al. Reversal of tumor immune inhibition using a chimeric cytokine receptor. Mol. Ther. 22, 1211–1220 (2014).
Mohammed, S. et al. Improving chimeric antigen receptor-modified T cell function by reversing the immunosuppressive tumor microenvironment of pancreatic cancer. Mol. Ther. 25, 249–258 (2017).
Wilkie, S. et al. Selective expansion of chimeric antigen receptor-targeted T-cells with potent effector function using interleukin-4. J. Biol. Chem. 285, 25538–25544 (2010).
Chang, Z. L. & Chen, Y. Y. CARs: synthetic immunoreceptors for cancer therapy and beyond. Trends Mol. Med. 23, 430–450 (2017).
Schwarz, K. A., Daringer, N. M., Dolberg, T. B. & Leonard, J. N. Rewiring human cellular input-output using modular extracellular sensors. Nat. Chem. Biol. 13, 202–209 (2017).
Baeumler, T. A., Ahmed, A. A. & Fulga, T. A. Engineering synthetic signaling pathways with programmable dcas9-based chimeric receptors. Cell Rep. 20, 2639–2653 (2017).
Scheller, L., Strittmatter, T., Fuchs, D., Bojar, D. & Fussenegger, M. Generalized extracellular molecule sensor platform for programming cellular behavior. Nat. Chem. Biol. 14, 723 (2018).
Green, M. R. & Sambrook, J. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Press, 2012).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Tiscornia, G., Singer, O. & Verma, I. M. Production and purification of lentiviral vectors. Nat. Protoc. 1, 241–245 (2006).
Liu, L. et al. Inclusion of Strep-tag II in design of antigen receptors for T-cell immunotherapy. Nat. Biotechnol. 34, 430–434 (2016).
Ghassemi, S. et al. Reducing ex vivo culture improves the antileukemic activity of chimeric antigen receptor (CAR) T cells. Cancer Immunol. Res. 6, 1100–1109 (2018).
Hollyman, D. et al. Manufacturing validation of biologically functional T cells targeted to CD19 antigen for autologous adoptive cell therapy. J. Immunother. 32, 169–180 (2009).
Vormittag, P., Gunn, R., Ghorashian, S. & Veraitch, F. S. A guide to manufacturing CAR T cell therapies. Curr. Opin. Biotechnol. 53, 164–181 (2018).
Chang, Z. L. et al. Rewiring T-cell signaling responses to extracellular soluble cues with chimeric antigen receptors. Mol. Ther. 27, abstr. 832 (2019).
Zah, E., Lin, M.-Y., Silva-Benedict, A., Jensen, M. C. & Chen, Y. Y. T cells expressing CD19/CD20 bispecific chimeric antigen receptors prevent antigen escape by malignant B cells. Cancer Immunol. Res. 4, 498–508 (2016).
Caruso, H. G. et al. Tuning sensitivity of CAR to EGFR density limits recognition of normal tissue while maintaining potent antitumor activity. Cancer Res. 75, 3505–3518 (2015).
Liu, X. et al. Affinity-tuned ErbB2 or EGFR chimeric antigen receptor T cells exhibit an increased therapeutic index against tumors in mice. Cancer Res. 75, 3596–3607 (2015).
Wang, X. et al. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 118, 1255–1263 (2011).
Wei, P. et al. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature 488, 384–388 (2012).
Wang, D. et al. A requirement for CARMA1 in TCR-induced NF-κB activation. Nat. Immunol. 3, 830–835 (2002).
Long, A. H. et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 21, 581–590 (2015).
Frigault, M. J. et al. Identification of chimeric antigen receptors that mediate constitutive or inducible proliferation of T cells. Cancer Immunol. Res. 3, 356–367 (2015).
Aivazian, D. & Stern, L. J. Phosphorylation of T cell receptor ζ is regulated by a lipid dependent folding transition. Nat. Struct. Biol. 7, 1023–1026 (2000).
Zhang, H., Cordoba, S.-P., Dushek, O. & van der Merwe, P. A. Basic residues in the T-cell receptor ζ cytoplasmic domain mediate membrane association and modulate signaling. Proc. Natl Acad. Sci. USA 108, 19323–19328 (2011).
Dobbins, J. et al. Binding of the cytoplasmic domain of CD28 to the plasma membrane inhibits Lck recruitment and signaling. Sci. Signal. 9, ra75–ra75 (2016).
Acknowledgements
Several protocols presented reflect discussion and contributions from present and past members of the Chen lab, notably E. Zah, P. Ho, M.-y. Lin, and M. Lorenzini, as well as members of the M. Jensen lab (Seattle Children’s Research Institute). The NFAT reporter Jurkat T-cell line was a gift from A. Weiss (University of California, San Francisco). The NFκB reporter Jurkat T-cell line was a gift from X. Lin (University of Texas MD Anderson Cancer Center). This work was supported by the National Institutes of Health (grant DP5OD012133, UC CAI grant U54HL119893, and UCLA CTSI grant UL1TR001881 to Y.Y.C.). Z.L.C. was supported by an NIH F30 Fellowship (F30CA183528). A.J.H. was supported by the NIH Biotechnology Training in Biomedical Sciences and Engineering Program (T32 GM067555).
Author information
Authors and Affiliations
Contributions
Z.L.C., A.J.H., and Y.Y.C. conceptualized and wrote the manuscript. Z.L.C. and Y.Y.C. developed the protocol, with additional contributions as reflected in the Acknowledgements section.
Corresponding author
Ethics declarations
Competing interests
Y.Y.C. and Z.L.C. declare competing interests in the form of a patent application whose value may be affected by the publication of this work.
Additional information
Peer review information Nature Protocols thanks Carl June, Atsushi Okuma and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Chang, Z. L. et al. Nat. Chem. Biol. 14, 317–324 (2018): https://doi.org/10.1038/nchembio.2565
Hou, A. J., Chang, Z. L., Lorenzini, M. H., Zah, E. & Chen, Y. Y. Bioeng. Transl. Med. 3, 75–86 (2018): https://doi.org/10.1002/btm2.10097
Integrated supplementary information
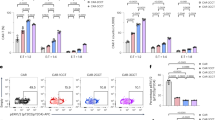
Supplementary Fig. 1 Comparison of quantifying cytokine secreted into supernatant and intracellular cytokine staining.
CD4 TGF-β CAR-T cells were stimulated with 5 ng/mL TGF-β for 24 hours. Side-by-side samples were prepared for analysis of supernatant or intracellular cytokine staining. a, Quantification of IFN-γ and TNF-α levels secreted into supernatant using a cytometric bead array showed comparable IFN-γ and TNF-α production. Data points from n = 3 (without TGF-β) or n = 2 (with TGF-β) samples are shown with means ± 1 standard deviation or total ranges, respectively. b, In contrast, intracellular cytokine staining detected more robust TNF-α production than IFN-γ production, indicating lower sensitivity for IFN-γ staining using the protocol described.
Supplementary Fig. 2 Cell sorting results are impacted by transduction efficiency of starting material.
Cells lines A and B were generated by transduction with a synthetic construct linked via a T2A “self-cleaving” peptide to truncated EGFR (EGFRt). The poorly transduced cell line (Cell Line A) and well-transduced cell line (Cell Line B) were then sorted according to steps 34-41. The poorly transduced starting material yielded an enriched product that was only 68.5% pure, while the well-transduced cell line yielded a 97.1% pure population.
Supplementary Fig. 3 Gating strategy for flow cytometry experiments.
a, An example gating scheme for all flow cytometry experiments is shown. Initial FSC-Area/SSC-Area gates are drawn to remove debris and dead cells. A subsequent FSC-Area/FSC-Height gate demarcates singlet events. b-d, Histograms are shown for flow cytometry of TGF-β CAR-expressing cell lines, gated for viable singlets, demonstrating upregulation of (b) NFAT-driven EGFP, (c) NFκB-driven EGFP, and (d) CD69. Plots in b-d are representative of data used to generate the bar graphs in Fig. 4a–c. b–d adapted from ref. 6, Springer Nature.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3.
Supplementary Data 1
Supplementary Spreadsheet
Rights and permissions
About this article
Cite this article
Chang, Z.L., Hou, A.J. & Chen, Y.Y. Engineering primary T cells with chimeric antigen receptors for rewired responses to soluble ligands. Nat Protoc 15, 1507–1524 (2020). https://doi.org/10.1038/s41596-020-0294-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0294-8
This article is cited by
-
Mesenchymal stromal cells with chimaeric antigen receptors for enhanced immunosuppression
Nature Biomedical Engineering (2024)
-
Non-invasive activation of intratumoural gene editing for improved adoptive T-cell therapy in solid tumours
Nature Nanotechnology (2023)
-
CARs: a new approach for the treatment of autoimmune diseases
Science China Life Sciences (2023)
-
Navigating CAR-T cells through the solid-tumour microenvironment
Nature Reviews Drug Discovery (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.