Abstract
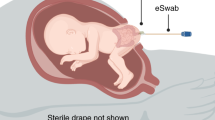
Mucosal immunity develops in the human fetal intestine by 11–14 weeks of gestation, yet whether viable microbes exist in utero and interact with the intestinal immune system is unknown. Bacteria-like morphology was identified in pockets of human fetal meconium at mid-gestation by scanning electron microscopy (n = 4), and a sparse bacterial signal was detected by 16S rRNA sequencing (n = 40 of 50) compared to environmental controls (n = 87). Eighteen taxa were enriched in fetal meconium, with Micrococcaceae (n = 9) and Lactobacillus (n = 6) the most abundant. Fetal intestines dominated by Micrococcaceae exhibited distinct patterns of T cell composition and epithelial transcription. Fetal Micrococcus luteus, isolated only in the presence of monocytes, grew on placental hormones, remained viable within antigen presenting cells, limited inflammation ex vivo and possessed genomic features linked with survival in the fetus. Thus, viable bacteria are highly limited in the fetal intestine at mid-gestation, although strains with immunomodulatory capacity are detected in subsets of specimens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All sequencing data associated with this study have been made publicly available. The 16S rRNA bacterial profiling data generated in this study are available in the EMBLI-EBI European Nucleotide Archive repository accession PRJEB25779 (https://www.ebi.ac.uk/ena). De novo assembled genomes were deposited at DDBJ/ENA/GenBank under accession number VFQL00000000 for Micro36. The genome version described in this paper is version VFQL01000000 for Micro36. Raw sequence reads used for genome assembly were deposited in NCBI Sequence Read Archive under BioProject accession PRJNA498337 for Micro36. RNA sequencing data are available in NCBI under PRJNA506292 accession. All additional datasets and materials are available from the corresponding author upon request; requests are promptly reviewed by the UCSF Office of Technology Management to verify whether the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a Material Transfer Agreement. Filtered and unfiltered OTU tables as well as metadata are provided as Supplementary Table 2 to this manuscript. Source data are provided for all figures and extended data.
References
Spencer, J., Dillon, S. B., Isaacson, P. G. & Macdonald, T. T. T cell subclasses in fetal ileum. Clin. Exp. Immunol. 65, 553–558 (1986).
Howie, D. et al. Extrathymic T cell differentiation in the human intestine early in life. J. Immunol. 161, 5862–5872 (1998).
McGovern, N. et al. Human fetal dendritic cells promote prenatal T cell immune suppression through arginase-2. Nature 546, 662–666 (2017).
Spencer, J., MacDonald, T. T., Finn, T. & Isaacson, P. G. The development of gut associated lymphoid tissue in the terminal ileum of fetal human intestine. Clin. Exp. Immunol. 64, 536–543 (1986).
Halkias, J. et al. CD161 contributes to prenatal immune suppression of IFN-γ-producing PLZF+ T cells. J. Clin. Invest. 130, 3562–3577 (2019).
Schreurs, R. R. C. E. et al. Human fetal TNF-α-cytokine-producing CD4+ effector memory T cells promote intestinal development and mediate inflammation early in life. Immunity 50, 462–476 (2019).
Li, N. et al. T cells are generated in the human fetal intestine. Nat. Immunol. 20, 301–312 (2019).
Aagaard, K. et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra65 (2014).
Collado, M. C., Rautava, S., Aakko, J., Isolauri, E. & Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 6, 23129 (2016).
Steel, J. H. et al. Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr. Res. 57, 404–411 (2005).
Lim, E. S., Rodriguez, C. & Holtz, L. R. Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome 6, 87 (2018).
Lauder, A. P. et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiot. Microbiome 4, 29 (2016).
de Goffau, M. C. et al. Human placenta has no microbiome but can contain potential pathogens. Nature 572, 329–334 (2019).
Chu, D. M. et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326 13 (2017).
Durack, J. et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat. Commun. 9, 707 (2018).
Yu, J. et al. Maternal exposure to farming environment protects offspring against allergic diseases by modulating the neonatal TLR–Tregs–Th axis. Clin. Transl. Allergy 8, 1–13 (2018).
Fujimura, K. E. et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191 (2016).
Gu, W. et al. Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high- abundance species in sequencing libraries and molecular counting applications. Genome Biol. 17, 41 (2016).
Trotter, A., Maier, L., Grill, H.-J., Wudy, S. A. & Pohlandt, F. 17β-estradiol and progesterone supplementation in extremely low-birth-weight infants. Pediatr. Res. 45, 489–493 (1999).
Yotis, W. & Stanke, R. Bacteriostatic action of progesterone on staphylococci and other microorganisms. J. Bacteriol. 92, 1285–1289 (1966).
Varghese, N. J. et al. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 43, 6761–6771 (2015).
Benach, J. et al. Structure of bacterial 3β/17β-hydroxysteroid dehydrogenase at 1.2 Å resolution: a model for multiple steroid recognition. Biochemistry 41, 14659–14668 (2002).
Hillas, P. J., del Alba, F. S., Oyarzabal, J., Wilks, A. & Ortiz De Montellano, P. R. The AhpC and AhpD antioxidant defense system of Mycobacterium tuberculosis. J. Biol. Chem. 275, 18801–18809 (2000).
Dons, L. E. et al. Role of the Listeria monocytogenes 2-Cys peroxiredoxin homologue in protection against oxidative and nitrosative stress and in virulence. Pathog. Dis. 70, 70–74 (2014).
Arrieta, M.-C. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7, 307ra152–307ra152 (2015).
Ferretti, P. et al. Mother-to-Infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145.e5 (2018).
Yassour, M. et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 24, 146–154.e4 (2018).
Egea, L., Hirata, Y. & Kagnoff, M. F. GM-CSF: a role in immune and inflammatory reactions in the intestine. Expert Rev. Gastroenterol. Hepatol. 4, 723–731 (2010).
Krause, P. et al. IL-10-producing intestinal macrophages prevent excessive antibacterial innate immunity by limiting IL-23 synthesis. Nat. Commun. 6, 7055 (2015).
Denning, T. L., Wang, Y. C., Patel, S. R., Williams, I. R. & Pulendran, B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat. Immunol. 8, 1086–1094 (2007).
Rosen, D. B. et al. Functional consequences of interactions between human NKR-P1A and its ligand LLT1 expressed on activated dendritic cells and B cells. J. Immunol. 180, 6508–6517 (2008).
Minich, J. J. et al. Quantifying and understanding well-to-well contamination in microbiome research. mSystems 4, e00186–19 (2019).
Martín, R. et al. Characterization of indigenous vaginal lactobacilli from healthy women as probiotic candidates. Int. Microbiol. 11, 261–266 (2008).
Chen, C. et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 8, 875 (2017).
Mukamolova, G. V. et al. The rpf gene of Micrococcus luteus encodes an essential secreted growth factor. Mol. Microbiol. 46, 611–621 (2002).
Duerkop, B. A., Vaishnava, S. & Hooper, L. V. Immune responses to the microbiota at the intestinal mucosal surface. Immunity 31, 368–376 (2009).
Mold, J. E. et al. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science 330, 1695–1699 (2010).
Mold, J. E. et al. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science 322, 1562–1565 (2008).
Pastor-vargas, C. et al. Detection of major food allergens in amniotic fluid: initial allergenic encounter during pregnancy. Pediatr. Allergy Immunol. 27, 716–720 (2016).
Iwai, S. et al. The lung microbiome of Ugandan HIV-infected pneumonia patients is compositionally and functionally distinct from that of San Franciscan patients. PLoS ONE 9, e95726 (2014).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. 108, 4516–4522 (2011).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Caporaso, J. G. et al. Correspondence QIIME allows analysis of high-throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nat. Publ. Gr. 7, 335–336 (2010).
Edgar, R. C. & Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482 (2015).
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Johansson, M. E. V. & Hansson, G. C. in Mucins: Methods and Protocols (eds McGuckin, M. A. & Thornton, D. J.) 229–235 (Humana Press, 2012).
Vaishnava, S. et al. The antibacterial lectin RegIII. Science 334, 255–258 (2011).
Weisburg, W. G., Barns, S. M., Pelletier, D. A. & Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 (1991).
Pruesse, E., Peplies, J. & Glöckner, F. O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Eren, A. M. et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, e1319 (2015).
Pritchard, L., Glover, R. H., Humphris, S., Elphinstone, J. G. & Toth, I. K. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods 8, 12–24 (2016).
Rinke, C. et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013).
Edgar, R. C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
Wood, D. E. & Salzberg, S. L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014).
Sekar, S., Mahadevan, S., Kumar, S. S. D. & Mandal, A. B. Thermokinetic responses of the metabolic activity of Staphylococcus lentus cultivated in a glucose limited mineral salt medium. J. Therm. Anal. Calorim. 104, 149–155 (2011).
Subashchandrabose, S., Smith, S. N., Spurbeck, R. R., Kole, M. M. & Mobley, H. L. T. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog. 9, 1–15 (2013).
Ducsay, C. A. et al. Gestational hypoxia and developmental plasticity. Physiol. Rev. 98, 1241–1334 (2018).
Oksanen, J. et al. Vegan: Community Ecology Package. R package v.2.0-10. (The R Project, 2013).
Kuznetsova, A., Brockhoff, P. B. & Christensen, R. H. B. lmerTest package: tests in linear mixed effects models. J. Stat. Softw. 82, 1–26 (2017).
Sprouffske, K. & Wagner, A. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinf. 17, 17–20 (2016).
Acknowledgements
We thank the tissue donors. We also thank M. Ng, H. Boushey and A. Molofsky for thoughtful critique of this manuscript. UCSF flow core provided instrumentation assistance, which was supported by National Institutes of Health (NIH) grant no. P30 DK063720 and NIH Shared Instrument grant no. 1S10OD021822-01. The FE-SEM and supporting facilities were obtained under National Science Foundation (NSF)-MRI award no. 0821619 and NSF-EAR award no. 0949176, respectively. This study was supported in part by the UCSF Clinical and Translational Science Institute Pilot Award for Basic and Translational Investigators grant no. 2014908 to J.H. E.R. was supported by NSF Graduate Research Fellowship grant no. 1650113 and by the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH F31 AI136336. J.H. was supported by NIH NIAID grant no. K08 AI128007. S.V.L. was supported by NIH NIAID grant no. AI114271 and AI113916. We thank C. Domingo for support of the electron microscopy core facility at San Francisco State University and D. Mars for assistance with FE-SEM sample preparation. We thank J. L. DeRisi at UCSF for generously sharing custom-made T7 RNA polymerase with our group. We thank E. Stapleton for illustration assistance and J. M. Rackaitis for support during these studies. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSF or the NIH.
Author information
Authors and Affiliations
Contributions
E.R. designed the study, performed research, analyzed the data and wrote the manuscript; J.H. contributed to study design, data analysis and manuscript development; E.M.F. assisted in bacterial identification, performed growth curve analysis and analyzed the data; C.H. contributed to study design and performed electron microscopy; V.F.M. performed immune cell isolations; E.D.C. assisted in sequencing methods development; K.E.F. contributed to data analysis and manuscript development; T.D.B. contributed to study design; S.V.L. designed the study, contributed to data analysis and wrote the manuscript. All authors discussed the results and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.V.L. is co-founder of Siolta Therapeutics Inc. and serves as both a consultant and a member of its Board of Directors. The Regents of the UCSF have filed a patent application (PCT/US2019/045354) on behalf of S.V.L. and E.R. relating to the methods and compositions of fetal bacteria.
Additional information
Peer review information Saheli Sadanand was the primary editor on this article and managed its editorial process in collaboration with the rest of the editorial team
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Low-burden bacterial signal in fetal meconium.
a. Fluorescent in situ hybridization probes targeting eubacteria (EUB) or non-targeting probe (NEUB) in 5 μm cryosections of human fetal (top panel) or murine (bottom panel) terminal ileum at 400x magnification. Arrowheads indicate EUB-positive findings in fetal sections. Scale bar corresponds to 50 mm. Quantification of independent fields of view (FOV) per mm within a representative b. human fetal (n = 28) or c. murine (n = 13) intestinal segments, where n is an independent segment of the intestine. For a-b, representative images of three fetal specimens and three mice. d. Total 16S copy number per 100 ng gDNA in meconium from mid-section of the fetal small intestine (n = 13), fetal kidney (n = 11), and procedural (n = 11), air (n = 8), or blank (n = 3) swab was quantified by qPCR of DNA extracts using a standard curve where n represents biologically independent samples; two-sided Satterthwaite’s method on linear mixed effects model to test for significance. e. Total 16S copy number per gram frozen sample in meconium from proximal (n = 12), mid (n = 13), and distal (n = 13) sections of the fetal small intestine where n represents independent biological samples across the length of the intestine or extraction buffer (n = 5, n represents biologically independent samples) was quantified by qPCR of DNA extracts using a standard curve; two-sided Wilcoxon rank sum test for significance compared to buffer control. Boxplots indicate the median (center), the 25th and 75th percentiles, and the smallest and largest values within 1.5 × the interquartile range (whiskers).
Extended Data Fig. 2 Depletion of mtDNA by Cas9 does not alter bacterial composition after 30 cycles of amplification.
16S rRNA V4 profiling of a subset (n = 10) of banked fetal meconium samples using different library preparation methods: gel extraction and 30 or 35 cycles of amplification, or 30 cycles combined with DASH performed on individual samples (Individual DASH) or on the library pool (Pooled DASH). a. Expansion in Enterobacteriaceae family is detected in 35-cycle amplification method, while small expansion of Pseudomonadaceae is detected post-DASH. Principal coordinates analysis of Bray Curtis distances of libraries using b. 30 cycles of amplification or c. 30 and 35 cycles of amplification, latter to provide an outgroup known to skew bacterial composition. Ellipses indicate 95% confidence intervals. All p-values were calculated using two-sided Satterthwaite’s method on Linear Mixed Effects (LME) modeling to correct for n = 10 paired samples that underwent multiple library preparation methods.
Extended Data Fig. 3 Sparse bacterial signal distinct from background detected in fetal meconium.
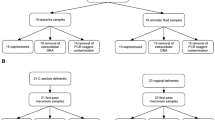
a. Histogram of number of operational taxonomic units (OTUs) per sample detected in fetal meconium from proximal-, mid-, or distal-segments of the small intestine after technical control filtering. b. Principal coordinates analysis (PCoA) of Bray Curtis distances of rareified bacterial profiles of proximal- (n = 32), mid- (n = 40), and distal- (n = 36) sections of the intestine. c. Inter- and intra-sample Bray Curtis distances between indicated comparisons of proximal- (n = 32), mid- (n = 40), and distal- (n = 36) intestinal sections. For a-b, n represents biologically independent fetal samples across the length of the intestine. d. Bacterial abundance ranks in fetal meconium, post-natal meconium, procedural swab, and kidney control. e. Relative abundance of select genera among samples dominated by OTU12, OTU10, or other OTUs. Symbols indicate samples with paired immunological datasets. PCoA of Bray Curtis distances of Lactobacillus-meconium (LM, n = 6), Micrococcaceae-meconium (MM, n = 9), or Other-meconium (OM, n = 25) compared to f. procedural swab (n = 14) or g. fetal kidney (n = 7) control, where n represents biologically independent fetal samples. h. Normalized read counts for Micrococcocaceae OTU10 in LM (n = 6), MM (n = 9), OM (n = 25), swab (n = 14), and fetal kidney (n = 7) control samples, where n represents biologically independent fetal samples; samples collected from the same fetus indicated by grey line, when possible. i. PCoA of Bray Curtis distances of unrareified and unfiltered bacterial profiles of meconium (n = 138 biologically independent fetal samples across three segments of the intestine) with technical negative controls (n = 48 biologically independent samples including extraction buffer, room air swab, pre-moistened swabs). LM (n = 6) and MM (n = 9) samples identified in later analyses are highlighted; significance was measured using two-sided Satterthwaite’s method on linear mixed effects model to test for significance and correct for repeated measures in b,h; ANOVA of linear mixed effects model was used to test for significance and correct for repeated measures in i, two-sided t-test was used for c, PERMANOVA was used in f-g. Boxplots indicate the median (center), the 25th and 75th percentiles, and the smallest and largest values within 1.5 × the interquartile range (whiskers).
Extended Data Fig. 4 Divergent epithelial transcriptome and lamina propria T cells in samples associated with LM, MM, or OM.
a. Proportion of PLZF+ CD161+ T cells among live, TCRβ+, Vα7.2−, CD4+ cells in intestinal lamina propria (LP, n = 28), mesenteric lymph node (MLN, n = 27), and spleen (SPL, n = 10) where n is a biologically independent fetal sample. b. Representative flow plots of mesenteric lymph node (top panel, gating control) or intestinal lamina propria (bottom panel) associated with MM and LM. Experiments were repeated 5 independent times for each LM and MM associated samples with similar results. c. Proportion of PLZF+ CD161+ T cells in intestinal lamina propria paired with LM, MM, or OM (LM-LP, n = 5; MM-LP, n = 5; OM-LP, n = 12) among live, TCRβ+, Vα7.2−, CD4+ cells. d. Principal components (PC) analysis of Euclidean distances of top 10,000 variable genes (by coefficient of variation) in LM associated epithelium (LM-E, n = 3), MM associated epithelium (MM-E, n = 7), or OM associated epithelium (OM-E, n = 3) as determined by RNA sequencing where n represents a biologically independent fetal sample. Kruskal-Wallis ANOVA, with Dunnet’s correction for multiple comparisons was used for a, c. PERMANOVA test for significance in d. Each dot represents a biological replicate. Boxplots indicate the median (center), the 25th and 75th percentiles, and the smallest and largest values within 1.5 × the interquartile range (whiskers).
Extended Data Fig. 5 Micrococcus fetal isolate exhibits high 16S rRNA V4 sequence identity to fetal meconium OTUs.
Alignment of 16S V4 rRNA gene sequences of Micro36 to OTU10. Percentage indicates identity to representative OTU sequence.
Extended Data Fig. 6 Fetal meconium Micrococcus isolate exhibits adaptation to the fetal environment.
Effects of 10−5 M progesterone (P4) and 10−6 M β-Estradiol (E2) on the growth a. MicroRef1 or b. MicroRef2 in carbon limiting media or of c. MicroRef1, d. MicroRef2, e. Micro36 with indicated concentrations of P4 and E2 or f. combinations of hormones compared to ethanol vehicle control, in carbon-rich media at 37 °C. Representative growth curves of three independent experiments measured by optical density at 600 nm (OD600), error bars denote standard error of the mean (SEM) between three technical experiments. For carbon-rich media conditions, integral of logistic regression model fitting was used to calculate area under the curve (auc) and change with respect to vehicle control is reported as Δauc. g. Intracellular survival of Micro36 or MicroRef1 or E. coli in RAW264.3 cells. ANOVA of generalized linear model of log(CFU + 1) against E. coli for each timepoint was used to calculate significance. Error bars indicate s.e.m. around center mean of n = 3 independent cell culture experiments. Growth of indicated strains on media with ( + ) or without (-) gentamicin (10μg mL−1) following 24-50 hours of intracellular growth in h. RAW264.7 cells or i. primary human fetal intestinal antigen presenting cells.
Extended Data Fig. 7 Genomic features of fetal Micrococcus isolate.
Alignment of all publicly available Micrococus genomes; single copy Micrococcus genes used for phylogeny (inset) and genes unique to Micro36 isolate are highlighted. Figure was generated using the Anvi’o package; each radial layer represents a genome; representative or reference genomes are colored in black indicated with asterisk; inner dendrogram represents hierarchical clustering of amino acid sequences based on their sequence composition and distribution across genomes; genomes are organized based on gene clusters they share using Euclidian distance and Ward ordination; outer ring represents single copy genes predicted using hidden markov model in Anvi’o package.
Extended Data Fig. 8 Prevalence of M. luteus in infants and mothers.
a. Percent identity of samples to 16S rRNA gene of Micro36 in three independent infant stool cohorts. Each symbol represents a sample with a positive hit ( > 97% sequence identity); symbol shape indicates cohort. Relative abundance of Micrococcus luteus in metagenomic sequencing cohorts across b. body sites at delivery in mother and infant within four months after birth, or c. in maternal stool around delivery and infant stool within the first three months of life. Metagenomic sequences obtained from two independent studies were classified using a custom kraken2 database including the fetal M. luteus Micro 36 genome. Correlation of gestational age with d. total number of OTUs or e. Micrococcaceae OTU10 count in mid-section meconium samples (n = 35 biologically independent fetal specimens) or f. among Micrococcaceae meconium (MM, n = 9 biologically independent fetal specimens). Pearson’s product-moment correlation coefficient and a one-sided t-distribution p-value is reported for d-f.
Extended Data Fig. 9 Fetal Micrococcus isolate promotes distinct APC and T cell phenotypes.
a. Proportion of live cells after treatment with media (n = 9) or Micrococcus (Micro36 n = 6, MicroRef1 n = 9, MicroRef2 n = 3) strains, where n represents biologically independent fetal specimens for the indicated treatment. ANOVA test for significance. b. HLA−DR+ CD45+ lin− cells pre- (left) and post- (right) fluorescence activated cell sorting (FACS). c. Proportion of naïve (CD45RA+ CCR7+), central memory (TCM, CD45RA− CCR7+), and effector memory T cells (TEM, CD45RA− CCR7−) among live, TCRβ+, CD4+ cells (left panel) and PLZF and CD161 expression among memory subsets, numbers indicate proportion in TEM (right panel). d. Pre- (left) and post- (right) FACS of effector memory T cells. e. Proportion of PLZF+ T cells or f. left, proportion of CD25hi FoxP3+ regulatory T cells (Tregs) and right, representative flow plots of FoxP3 and CD25 expression among intestinal live, TCRβ+, CD4+, Vα7.2−, cells after exposure to splenic APCs pretreated with media or Micrococcus (Micro36, MicroRef1) strains for n = 5 biologically independent fetal specimens. Concentration of g. IL-17A, h. IL-17F, i. GM-CSF, j. IL-4, k. IL-10, l. IL-13, m. TNFα in culture supernatants of lamina propria T cell co-cultures with splenic antigen presenting cells pre-exposed to media (n = 7) or Micrococcus (Micro36 n = 6, MicroRef1 n = 7, MicroRef2 n = 7) strains, where n represents biologically independent fetal specimens for the indicated treatment. For b-d, f numbers indicate mean proportion and standard error of the mean (s.e.m.) representative of five independent experiments. For e-f, g-m two-sided Satterthwaite’s method on linear mixed effects model was used to test for significance between strains, controlling for repeated measures of cell donor. Positive LME residuals are plotted for g-m. Each dot represents an independent fetal sample, unless otherwise indicated. Boxplots indicate the median (center), the 25th and 75th percentiles, and the smallest and largest values within 1.5 × the interquartile range (whiskers).
Supplementary information
Supplementary Information
Supplementary Figs. 1–3.
Supplementary Tables
Supplementary Tables 1–9.
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 9
Statistical Source Data
Rights and permissions
About this article
Cite this article
Rackaityte, E., Halkias, J., Fukui, E.M. et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med 26, 599–607 (2020). https://doi.org/10.1038/s41591-020-0761-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-020-0761-3
This article is cited by
-
The maternal gut microbiome in pregnancy: implications for the developing immune system
Nature Reviews Gastroenterology & Hepatology (2024)
-
Investigating prenatal and perinatal factors on meconium microbiota: a systematic review and cohort study
Pediatric Research (2024)
-
A genome-wide association study reveals the relationship between human genetic variation and the nasal microbiome
Communications Biology (2024)
-
A common trajectory of gut microbiome development during the first month in healthy neonates with limited inter-individual environmental variations
Scientific Reports (2024)
-
RNA-based amplicon sequencing is ineffective in measuring metabolic activity in environmental microbial communities
Microbiome (2023)