Abstract
The scarcity of donor organs may be addressed in the future by using pigs to grow humanized organs with lower potential for immunological rejection after transplantation in humans. Previous studies have demonstrated that interspecies complementation of rodent blastocysts lacking a developmental regulatory gene can generate xenogeneic pancreas and kidney1,2. However, such organs contain host endothelium, a source of immune rejection. We used gene editing and somatic cell nuclear transfer to engineer porcine embryos deficient in ETV2, a master regulator of hematoendothelial lineages3,4,5,6,7. ETV2-null pig embryos lacked hematoendothelial lineages and were embryonic lethal. Blastocyst complementation with wild-type porcine blastomeres generated viable chimeric embryos whose hematoendothelial cells were entirely donor-derived. ETV2-null blastocysts were injected with human induced pluripotent stem cells (hiPSCs) or hiPSCs overexpressing the antiapoptotic factor BCL2, transferred to synchronized gilts and analyzed between embryonic day 17 and embryonic day 18. In these embryos, all endothelial cells were of human origin.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The scRNA-seq data that support the findings of this study have been deposited in NCBI Sequence Read Archive (SRA) database under accession number PRJNA577880. All data will be made available by the authors upon reasonable request. All unique materials used in these studies are readily available from the authors or from commercial sources (Supplementary Tables 1 and 2). Gene-edited primary cell lines are limited in number but will be made available pending their supply.
References
Kobayashi, T. et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142, 787–799 (2010).
Usui, J. I. et al. Generation of kidney from pluripotent stem cells via blastocyst complementation. Am. J. Pathol. 180, 2417–2426 (2012).
Ferdous, A. et al. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc. Natl Acad. Sci. USA 106, 814–819 (2009).
Rasmussen, T. L. et al. ER71 directs mesodermal fate decisions during embryogenesis. Development 138, 4801–4812 (2011).
Koyano-Nakagawa, N. et al. Etv2 is expressed in the yolk sac hematopoietic and endothelial progenitors and regulates Lmo2 gene expression. Stem Cells 30, 1611–1623 (2012).
Rasmussen, T. L. et al. VEGF/Flk1 signaling cascade transactivates Etv2 gene expression. PLoS One 7, e50103 (2012).
Rasmussen, T. L. et al. Etv2 rescues Flk1 mutant embryoid bodies. Genesis 51, 471–480 (2013).
Wu, J. et al. Interspecies chimerism with mammalian pluripotent stem cells. Cell 168, 473–486 (2017).
Lee, D. et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell 2, 497–507 (2008).
Hadjantonakis, A. K., Macmaster, S. & Nagy, A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnology 2, 11 (2002).
Shi, X. et al. The transcription factor Mesp1 interacts with cAMP-responsive element binding protein 1 (Creb1) and coactivates Ets variant 2 (Etv2) gene expression. J. Biol. Chem. 290, 9614–9625 (2015).
Whitworth, K. M. et al. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol. Reprod. 91, 78 (2014).
Giraldo, A. M., Ball, S. & Bondioli, K. R. Production of transgenic and knockout pigs by somatic cell nuclear transfer. Methods Mol. Biol. 885, 105–123 (2012).
Wu, J. et al. Generation of human organs in pigs via interspecies blastocyst complementation. Reprod. Domest. Anim. 51, 18–24 (2016).
Yamaguchi, T. et al. Interspecies organogenesis generates autologous functional islets. Nature 542, 191–196 (2017).
Iacovino, M. et al. A conserved role for Hox paralog group 4 in regulation of hematopoietic progenitors. Stem Cells Dev. 18, 783–792 (2009).
Chen, G. et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8, 424–429 (2011).
Cerbini, T. et al. Transcription activator-like effector nuclease (TALEN)-mediated CLYBL targeting enables enhanced transgene expression and one-step generation of dual reporter human induced pluripotent stem cell (iPSC) and neural stem cell (NSC) lines. PloS one 10, e0116032 (2015).
Koyano-Nakagawa, N. et al. Feedback mechanisms regulate Ets Variant 2 (Etv2) gene expression and hematoendothelial lineages. J. Biol. Chem. 290, 28107–28119 (2015).
Lai, L. & Prather, R. S. Production of cloned pigs by using somatic cells as donors. Cloning Stem Cells 5, 233–241 (2003).
Sembon, S. et al. A simple method for producing tetraploid porcine parthenogenetic embryos. Theriogenology 76, 598–606 (2011).
Machaty, Z., Wang, W. H., Day, B. N. & Prather, R. S. Complete activation of porcine oocytes induced by the sulfhydryl reagent, thimerosal. Biol. Reprod. 57, 1123–1127 (1997).
Whitworth, K. M., Zhao, J., Spate, L. D., Li, R. & Prather, R. S. Scriptaid corrects gene expression of a few aberrantly reprogrammed transcripts in nuclear transfer pig blastocyst stage embryos. Cell. Reprogram. 13, 191–204 (2011).
Zhao, J. et al. Significant improvement in cloning efficiency of an inbred miniature pig by histone deacetylase inhibitor treatment after somatic cell nuclear transfer. Biol. Reprod. 81, 525–530 (2009).
Redel, B. K. et al. Glycine supplementation in vitro enhances porcine preimplantation embryo cell number and decreases apoptosis but does not lead to live births. Mol. Reprod. Dev. 83, 246–258 (2016).
Allard, J. et al. Immunohistochemical toolkit for tracking and quantifying xenotransplanted human stem cells. Regen. Med. 9, 437–452 (2014).
Nguyen, Q. H. et al. Single-cell RNA-seq of human induced pluripotent stem cells reveals cellular heterogeneity and cell state transitions between subpopulations. Genome Res. 28, 1053–1066 (2018).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Acknowledgements
This work was supported by a grant from the Department of Defense (grant no. 11763537 to D.J.G.) and Minnesota Regenerative Medicine (to D.J.G.). We thank J. Dutton (University of Minnesota) and G. Daley (Harvard Medical School) for providing hiPSC lines. We thank C. Chapman for assisting with the hiPSC culture studies, E. Skie for genetic analyses, Y. Ren and C. Walter for the FACS analysis, and B. Coffin, A. Arnason, L. Meisner and D. Ly for morphological analyses. We thank A. Caplan for critical comments during the course of these studies. We acknowledge the Mouse Genetics Laboratory, Veterinary Diagnostic Laboratory and the University Imaging Center at the University of Minnesota, NorthStar Genomics, Recombinetics, DeSoto Biosciences, grafikalabs (http://grafikalabs.com/) and MOFA Global for their technical assistance. We also thank R. Prather and the National Swine Resource and Research Center at the University of Missouri for providing technical training and assistance (U42 OD011140).
Author information
Authors and Affiliations
Contributions
N.K.-N., M.G.G. and D.J.G. conceived the project and S.D., N.K.-N., D.Y., M.G.G. and D.J.G. wrote the manuscript. S.D., N.K.-N., T.R., O.G., S.K., B.N.S., G.M., X.P., K.-D.C., P.P., W.G., J.H.H., D.M. and C.V.W. designed and performed experiments, and analyzed the data. N.K.-N., M.G.G. and D.J.G. supervised the project. All authors commented on and edited the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
D.J.G. and M.G.G. are co-founders of NorthStar Genomics.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Lineage complementation occurs in a cell-autonomous manner in Etv2 mutant hosts.
(a, b) ES/EB differentiation assay. The EYFP-transgenic wild type ES line 7AC5 was differentiated alone (EYFP), with WT ES cells (WT/EYFP), or with Etv2 knockout ES cells (KO/EYFP) in the ES/EB system. Percentage of EYFP positive cells within the endothelial (a; Flk1+/Tie2+ ; n = 3 biologically independent samples) and hematopoietic (b; CD41+; ; n = 4 biologically independent samples) populations. Statistical significance (P < 0.0001) determined by one-way ANOVA with a Bonferroni posttest for repeated measures. Exact P-values are mentioned in the figure panels. n.s. = non-significant (c) Diagram summarizing the results in a and b. (d–k) Blastocyst complementation of Etv2 mutant mouse embryos. Representative images and cross-sections of chimeric embryos with Etv2dKI/+ hemizygous host (d, f, h, j) and Etv2dKI/KO mutant host (e, g, i, k) (n = 3 for each group). Sections were immunostained with antibodies to GFP (f-i, green), Endomucin (f, g, j, k, red), or Tie2 (insets in g, i, k, red). Sections in f and g are enlarged in Supplementary Fig. 2a and Supplementary Fig. 2c, and the green channels are shown in Supplementary Fig. 2b and Supplementary Fig. 2c. da: dorsal aorta, ht: heart, nt: neural tube. Scale bars, 1mm (d, e), or 100 μm (f-k). (l-s). Representative FACS profiles (l, n, p, r) and quantification (m, o, q, s) of endothelial (l-o) and hematopoietic (p-s) lineages (n = 7 WT chimeric embryos, n = 9 dKI/KO chimeric embryos and n = 3 for each of the other groups). Flk1+ populations in l and CD41+ populations in p were analyzed for EYFP expression in n and r, respectively. Data represent mean ± SEM.
Supplementary Figure 2 Lineage complementation occurs in a cell-autonomous manner in Etv2 mutant hosts.
Sections in Supplementary Fig. 1f and Supplementary Fig. 1g are enlarged in a and c, and the green channels are shown in b and d. da: dorsal aorta, ht: heart, nt: neural tube. Bracket in b denotes examples of EYFP positive cells that are not localized to the endothelial lineage in the hemizygous host. Scale bar equals 100 μm.
Supplementary Figure 3 Schematic overview for the blastocyst complementation studies using the porcine model.
The figure demonstrates the flow of experiments that were performed, which began with the generation of ETV2 mutated porcine fibroblasts using CRISPR/Cas9 gene-editing technology. Somatic cell nuclear transfer (SCNT) technology was used to clone the ETV2 mutated nucleus from fibroblasts into the enucleated pig primary oocytes. The developing morulae from the cloned oocytes were injected with either porcine stem cells (blastomeres) for pig-pig or human hiPSCs for human-pig complementation studies. The chimeric complemented embryos were either cultured in vitro or transferred into synchronized gilts for in vivo studies. We performed proliferation and survival assays, immunohistochemistry, in situ hybridization, human-pig cell integration and communication studies and TUNEL assays in vitro. For in vivo studies, embryos were harvested at defined embryonic stages [E18, E24 or FT (full term)] and analyzed using FACS, gDNA and RNA qPCR, immunohistochemistry, in situ hybridization or methylcellulose assays.
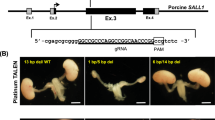
Supplementary Figure 4 Targeting and genotyping strategies for generation of ETV2 knockout fibroblasts.
(a) Diagram of porcine (sus scrofa) ETV2 gene. Red arrows indicate location of guide RNAs and black arrows indicate genotyping primers. (b) Genotyping PCR. Deletion was confirmed by the absence of 5’ (5F1/5R1), 3’ (3F1/3R1), and internal (internal F1/R1) amplifications, and the presence of 5F1/3R1 amplification. 600 clones were screened and three independent clones with biallelic deletion of ETV2 were obtained and shown. (c) Expected fragment sizes for genotyping PCR are indicated.
Supplementary Figure 5 Gating strategy used for FACS analyses.
For the analyses of flow cytometry experiments, the gating of acquired events was based on the forward scatter (FSC-A) and side scatter (SSC-A). Live cells were gated based on the propidium iodide (PI) negative and side scatter (SSC-A) plots. Singlets were gated based on the height and width of FSC (FSC-H vs. FSC-W) and SSC (SSC-H vs. SSC-W) for their cell size and granularity. Single cells stained with fluorescence-labeled antibodies specific for cell surface markers were analyzed and the data which were obtained were processed using FlowJo Ver10.4.2 software.
Supplementary Figure 6 Porcine blastomeres integrate into parthenogenetic embryos and develop with the host.
(a) Schematic outline of the experimental scheme. Parthenogenetic embryos were generated by electrical stimulation of matured oocytes and cultured. At day 4, embryos reached the morula stage and were injected with 2–8 blastomeres expressing GFP. (b) Representative images of GFP-labeled blastomeres developing with the host parthenogenote. (c) Number of parthenogenotes containing GFP-labeled cells. The bar graph shows results obtained from three biologically independent (n = 3) experiments. (d) Number of GFP+ cells per parthenogenote. 80 parthenogenotes (n = 80) were injected with GFP+ blastomeres on D4. Nine out of 15 biologically independent parthenogenotes analyzed were positive for GFP expressing cells at D10. The p-value was determined by one-way ANOVA and non-parametric method with Brown-Forsythe test. Data represent mean±SEM (e) Immunohisochemistry revealed that GFP+ cells integrated and contributed to both OCT4+/CDX2+ (open arrowheads) and OCT4+/CDX2- (filled arrowheads) populations. Data show representative images from n = 6 biologically independent samples.
Supplementary Figure 7 Complementation of the hematoendothelial lineages by GFP-labeled blastomeres in the ETV2 knockout embryos.
A total of 1,322 complemented embryos were transferred to 8 pseudopregnant gilts and 8 WT-ETV2 null porcine complemented embryos were analyzed at distinct embryonic stages (Also see Fig. 2). (a-c) Wholemount views of an E18 complemented embryo under brightfield (a) or GFP epifluorescence (b). (c) is a high magnification image of the allantois. (d-l) Cross sections were stained with antibodies to GFP (e-h, j-l), TIE2 (d, f), and SM22 (i). DAPI is used as a nuclear counterstain (d, e, h, i, j, k, l). Scale bars, 1mm (a, b), 100 μm (c), 50 μm (d-f, j-n), or 10 μm (g, h). (m) Percentage of GFP+ cells in different tissues in the complemented embryo. Endothelial (n = 632) and smooth muscle (n = 237) lineages were identified by immunohistochemistry. Mesenchyme (n = 1,219), Neural tube (n = 2,043) and Intestine (n = 668) were defined morphologically. Note that both embryonic and extraembryonic endothelial lineages are derived 100% from GFP+ cells, whereas other tissues are chimeric. Scale bars, 1mm (a, b), 100 μm (c), 50 μm (d-f, i-l), or 10 μm (g, h). Total number of GFP+cells analyzed include 4,799 cells. Data represent mean±SEM.
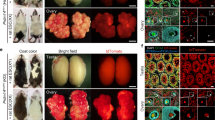
Supplementary Figure 8 Complementation of ETV2 knockout embryos (full term; n=3 biologically independent samples).
Body weights of all full term offspring were within range of normal birth weights (mean±SEM are 1.14±0.127 kg; n = 3). (a-k) Tissues were examined morphologically and then harvested for immunohistochemical analysis. a) The heart (open arrowhead) and descending aorta (arrowhead) were morphologically normal in the complemented animals. (b and c) The hearts were isolated and imaged to demonstrate the morphologically normal aortic arch in the complemented animals. (d and e) Wholemount images of the descending aorta demonstrate a longitudinal (d) section and cross sectional (e) image of the complemented aorta. Sections of skeletal muscle (f-j) and heart (j and k) were immunohistochemically stained for GFP (green) and the endothelial marker VWF (red). Note all the VWF endothelial cells were labeled with GFP. Scale bars for panels d-e = 2mm, panels f-g = 100μm, panels h-k = 50μm. (l) Quantification of chimeric rate by genomic PCR in indicated organs. ETV2 KO fibroblast and GFP Tg fibroblasts are parental cells used for cloning. Data shown are in triplicate for each tissue and plotted as mean±SEM..
Supplementary Figure 9 Evaluation of culture conditions for hiPSC-injected parthenotes.
(a) Development rate of parthenogenotes to morula (compaction) and blastocyst stages in three porcine media (n = 40 for each group). (b-g) Cell viability was assessed by TetraZ assay after culturing three hiPSC lines at indicated temperatures in NCSU-23 (b-d) or PZM-MU2 (e-g) media mixed with mTesRTM1 medium. Data shown are from n = 4 biologically independent experiments. (h) Number of porcine parthenogenotes containing live hiPSCs (shiPS9-1) over short-term culture (D4, n = 5; D5, n = 47; D6, n = 8 biologically independent samples). (i) Number of live hiPSC cells per parthenogenotes over short-term culture (D4, n = 5; D5, n = 47; D6, n = 8 biologically independent samples). Data represent mean±SEM. (j, k) Blastocyst rate of parthenogenotes injected with hiPSCs (SH91) at d4 or d6 after activation, scored at 24h (j) and 48h (k). Dot plots show results from n = 4-9 biologically independent experiments and plotted as mean±SEM.
Supplementary Figure 10 Detection of hiPSCs in porcine host.
(a-g) hiPSCs were prelabeled with DiI (a-c) or EdU (d-g) and injected into parthenogenotes. Embryos were analyzed 48hrs later. DiI and HNA immunohistochemistry (a-c) or EdU and HNA detection identified the same cells (d-f). Note that panel g is a brightfield image of panels d-f. (h-k) Genomic in situ hybridization using primate specific ALU probe (h,i) and HNA immunohistochemistry (j, k) on human (h, j) and pig (i, k) sections reveal specificity of detection. Scale bars, 50 μm. Data shown are representative images from n = 5 biologically independent samples. (l, m) Genomic qPCR using ALU and CYTB primers. Data shown are in triplicate for each dilution series and plotted as mean±SEM.
Supplementary Figure 11 Long-term culture of hiPSC-injected parthenogenotes.
Examples of hiPSCs proliferating in long-term culture of parthenogenotes (a). Number of parthenogenotes containing GFP+ hiPSCs (b). Number of GFP+ hiPSCs within each parthenogenote over time (c). For the long-term culture experiments (n = 64 parthenogenotes) GFP+cells were analyzed at D4-D10. Data represents mean±SEM.
Supplementary Figure 12 Generation of human iPS cell lines harboring copGFP and BCL2 knock in insertions.
(a) Schematic outlining the knock in strategies for copGFP to the CLYBL locus (top) and BCL2 over expression to the AAVS1 locus (bottom) (CF1/CR1;CF2/CR2). (b) PCR gel images showing correct targeting of RhiPSC11 cGFP in the CLYBL locus (5’CF1/CR1 & 3’CF2/CR2 CLYBL locus targeting validation) and RhiPSC11 cGFP-BCL2 both in the CLYBL and AAVS1 loci (5’AF1/AR1 & 3’AF2/AR2 AAVS1 locus targeting validation), yet not in RhiPSC11 WT nor NTC (no template control). Data shown are representative of two independent PCR verifications of the correctly targeted lines. (c) Immunoblot image showing over expression of BCL2 in RhiPSC11 cGFP-BCL2 and the positive control (PC) but not in the WT hiPSCs. Data shown are representative of two independent immunoblot verifications of the correctly targeted lines.
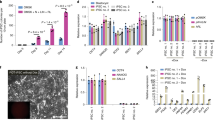
Supplementary Figure 13 BCL2 overexpression results in increased human:porcine chimerism.
(a) qPCR analysis was performed using genomic DNA from chimeric human:porcine embryos/embryo products at E17-E18 following delivery of hiPSCs (a) or BCL2-hiPSCs (b). Note the marked increase in efficiency using the BCL2-hiPSCs (compared to hiPSCs) in the top ten embryos. (c) and (d) Percentage of TIE2 positive cells among all the GFP positive cells and percentage of GFP positive cells among TIE2 positive cells, respectively in the human (BCL2-hiPSC):porcine chimeric embryos. (e) Comparison of Alu fold change using PCR in chimeric ETV2 null embryos that were complemented with either BCL2-hiPSCs (n = 63) or hiPSCs (n = 23) and transferred to surrogate gilts. The black dots represent individual samples in each group. The BCL2-hiPSC group emphasizes the increased efficiency of human-porcine chimerism compared to the hiPSCs group. The whisker plot shows all data points (minimum to maximum).
Supplementary information
Supplementary information
Supplementary Figs. 1–13 and Supplementary Tables 1–2.
Rights and permissions
About this article
Cite this article
Das, S., Koyano-Nakagawa, N., Gafni, O. et al. Generation of human endothelium in pig embryos deficient in ETV2. Nat Biotechnol 38, 297–302 (2020). https://doi.org/10.1038/s41587-019-0373-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0373-y
This article is cited by
-
ETV2/ER71, the key factor leading the paths to vascular regeneration and angiogenic reprogramming
Stem Cell Research & Therapy (2023)
-
Humanizing pig kidneys via chimeric complementation
Cell Research (2023)
-
A 3-Gene Random Forest Model to Diagnose Non-obstructive Azoospermia Based on Transcription Factor-Related Henes
Reproductive Sciences (2023)
-
Why it is important to study human–monkey embryonic chimeras in a dish
Nature Methods (2022)
-
New concepts for generating interspecies chimeras using human pluripotent stem cells
Protein & Cell (2022)