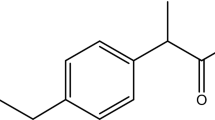
Abstract
Background
In this study, the biodegradation of N-acetyl-para-aminophenol also known as acetaminophen (APAP, paracetamol) was studied by bacterial strain Bacillus drentensis strain S1 (accession no. KY623719) isolated from sewage sample.
Results
The Bacillus drentensis strain S1 was isolated from the sewage sample using the enrichment culture method. As per our knowledge this is the first Bacillus drentensis strain reported for the degradation of APAP. In this study a 20-L batch reactor was employed for degradation of APAP. The maximum specific growth rate (μmax) was observed at 400 mg/L concentration of APAP. The pilot-scale anaerobic batch reactor of was stable and self-buffered. The degradation in pilot-scale reactor was slow as compared to batch experiments due to fluctuation in pH and exhaustion of nutrients. Design-Expert® software was used for optimization of conditions for APAP degradation; such as temperature (40 °C), pH (7.0), concentration of APAP (300 g/L) and agitation speed (165 rpm). The FTIR and GC–MS were used to identify the degradation metabolites. The intermediates of degradation like 2-isopropyl-5-methylcyclohexanone and phenothiazine were observed, based on these results the metabolic pathway has been predicted.
Conclusions
The optimization, kinetic, batch study and pilot study indicates the potential of Bacillus drentensis strain S1 for degradation of acetaminophen. The experimental design, optimization and statistical analysis were performed by Design Expert® software. The optimal growth condition for Bacillus drentensis strain S1 was found to be at temperature 40 °C, pH 7, acetaminophen at concentration of 300 (mg/L) and agitation speed 165 rpm. The GC–MS and FTIR was used for identification of metabolites produced during acetaminophen degradation and the partial metabolic pathway for degradation of acetaminophen was also proposed

.
Similar content being viewed by others
Background
The pharmaceuticals pollutants viz. analgesics, anti-inflammatory drugs and non-steroidal anti-inflammatory drugs (NSAIDs), psychiatric drugs, β-blockers, other illicit drugs, etc., are classified as emerging organic contaminants (EOCs). They are detected in rivers, lakes, ground water, marine and various aquatic ecosystems (Wu et al. 2012). These compounds are designed to cure diseases in humans and animals. Presently, many of these compounds are detected worldwide ranging from ng/L to μg/L. The concentration of these compounds is increasing day by day, due to high rates of production and consumption. This has lead to adverse effects on ecosystem due to their biologically active nature (Onesios et al. 2009). One of the most frequently detected pharmaceutical and personal care products (PPCP) in treated and untreated waste water is acetaminophen (APAP), commonly known as paracetamol and chemically N-acetyl-para-aminophenol. This is the one of most salable over the counter medicines, highly produced and prescribed. APAP is prescribed as fever reducer, pain relief, analgesic and antipyretic drug (Chandrasekharan et al. 2002). The metabolism of APAP occurs in the liver of patient, and it is not completely metabolized and secreted during excretion. The concentration of APAP as excretion varies from person to person and the average concentration lies between 60 and 70% (Khetan and Collins 2007). The APAP has chemical properties, like high solubility and hydrophilic nature, which enables the entry of this compound in the aquatic environment. Also this is one of the most identified PPCPs, detected in water resources around the globe. Their entry into the environment leads to production of intermediates metabolites and these metabolites are highly toxic compared to parent molecule and having more adverse effect in the environment (Balakrishna et al. 2017).
APAP is more frequently detected in waste water treatment plants (WTPs) commonly used for treatment of domestic waste water. The concentration of APAP in these WTPs is 40 times higher in developing countries like India due to high population (Balakrishna et al. 2017). The presence of APAP and its metabolites in drinking water resources raised issues related to human health. Presently, there is no such evidence available, which has shown the adverse effects of dissolved APAP on human health (Minto et al. 1997). But the high therapeutic use of the APAP can lead to nephrotoxicity and teratogenic effects. The use of high concentration of APAP is reported to cause adverse effect on human liver (Khan, et al. 2006). Chlorination is most commonly used method for drinking water treatment in many developing countries. A higher emission and mass load of PPCPs has been reported in India (Subedi et al. 2017). During chlorination, the APAP is transformed into various intermediates, such as N-acetyl-p-benzoquinoneimine (NAPQI) and 1,4-benzoquinone. These transformed compounds having higher toxicity compared to APAP (Bedner and MacCrehan 2006). Now a days, the WTPs in many countries use advanced methods to remove such compounds such as electrochemical (Brillas et al. 2005, Waterston et al. 2006); ozonation; H2O2/UV; H2O2/Fe2+/UV oxidation of waste water (Skoumal et al. 2006; Vogna et al. 2002) and semiconductor photo-catalysis methods. These technologies having high operation cost while in some methods APAP gets transformed into a more toxic form of intermediate compounds (Yang et al. 2008, 2009).
As compared to above-mentioned methods, biodegradation is considered as the eco-friendly and cost-effective option. With the help of biodegradation mechanism, APAP is degraded and converted into low molecular weight dead-end products (Hasan et al. 2012; Chen et al. 2010). The Penicillium sp. transforms APAP into 4-aminophenol one of the dead-end metabolite and acetate (Hart and Orr 1975). The Rhodococcus strain degrades APAP into intermediates compounds: 4-aminophenol, hydroquinone and catechol (Ismail et al. 2017). Bart et al. (2011) purposed the metabolic pathway for the degradation of APAP by D. tsuruhatensis and P. aeruginosa and it was observed that hydroquinone as an intermediate produced during degradation. Strains like Burkholderia sp. AK-4 have the capability to degrade aminophenol into 1,2,4-trihydroxybenzene with the intermediate of 1,4-hydroxybenzene (Zhang et al. 2013). The biodegradation of APAP by soil microorganisms occurs by the cleavage of aromatic ring of APAP into 3-hydroxyacetaminophen and N-acetyl benzoquinoneimine and this occurs by the mechanisms involving hydroxylation, methylation and oxygenation reactions (Li et al. 2014). The bacterium plays a vital role in the APAP degradation and contributes to eco-friendly environment (Hu et al. 2013). In this study, the APAP-degrading bacterium Bacillus drentensis strain S1 was isolated by enrichment culture method from the sample collected from highly polluted sewage. The strain was identified by microscopy, biochemical, and molecular characterization-based methods. The biodegradation mediated by the Bacillus drentensis strain S1 was performed in 250-mL flasks and also in 20-L pilot-scale reactor. The metabolites produced during degradation were analyzed by FTIR and GC–MS-based methods.
Materials and methods
Chemicals used and collection of samples
Acetaminophen (99% purity) was purchased from Sigma Aldrich (USA). Other chemicals used in this study were purchased from HiMedia (Mumbai, India). The sewage samples used in this study were collected from waste water drain in Sonipat, Haryana, India (28.98°N 77.02°E). This sample contains non-treated waste water mainly from household waste and small-scale industrial discharges. These samples contain microbial consortium having ability to degrade different compounds present in waste water. They were presumed to be the best source for showing adaptation of microorganisms in it and compounds gets transformed by the metabolic activity of microorganisms. The samples were transported to the laboratory in a refrigerated container and stored at 4 °C. The samples were settled down for heavy particles and the sediment was discarded before use. Later, the samples were filtered just before use through Whatman filter paper (90 mm pore size) to remove any suspended particles.
Enrichment, isolation and screening of APAP-degrading bacteria
APAP biodegradation tests were performed in 250-mL conical flasks (abiotic tests) supplemented with Bushnell Haas Medium (BHM, HiMedia, Mumbai, India), APAP and waste water sample. The flasks were incubated at 37 °C in rotary shaker at 150 rpm according to general standard conditions (including a blank test without activated waste water sample and an adsorption test by using autoclaved activated sample, etc.). The degradation was checked after at regular intervals of 8 h. Consequently, 1 mL of the mixture containing APAP-degrading strain was analyzed for pH and APAP concentration. The mixture was suspended into fresh sterilized media having APAP (100–500 mg/L), concentration increased in stepwise manner. After, 5 days the disposable plastic plates contain MSM agar medium supplemented with 100 to 500 mg/L of APAP (this also includes a blank test and an adsorption test). The strain isolation was performed by enrichment culture technique using liquid MSM (pH of 7.2 ± 0.5) in 250 mL flask as per the method prescribed by Jameson (1961). These plates were incubated in aerobic condition at 37 °C in a BOD incubator. After the incubation of 24 h, the bacterial colonies having different morphological features were sub-cultured on nutrient agar plates.
Experimental design, degradation optimization studies and statistical analysis
The effect on various parameters viz. temperature, pH, shaking speed and concentration was studied for the degradation of APAP mediated by the addition of culture of S1 strain. These experiments were performed in 100-mL flasks containing varying concentrations of MSM, APAP and biomass. Design-Expert® software (version DX 6.0.1, Stat-Ease, Minneapolis, 2005) was used for designing the experiment, for the best fit value of physical parameters and thereby statistical analysis performed. Box–Behnken design (BBD) (Mirizadeh et al. 2014) with quadratic model was used to identify the combined effect of four independent variables with the range as parameters viz. temperature (20–60 °C), pH (5–9), agitation speed (80–250 rpm) and concentration of APAP (100–500 mg/L). BBD-proposed three-level designs were used for fitting response surfaces to get the best values for degradation of APAP obtained for different variables based on second-order polynomial model. Each factor was coded at three levels and 29 experiments were performed in shaking-flask set at previously fixed conditions. Thereafter, statistically significant values were analyzed by Design-Expert® software. The response surface methodology (RSM) plots were used to identify the parameters for optimization of specific growth conditions.
The analysis of variance (ANOVA) and multiple regression analysis were used to know the best fit response by using the quadric model in Box–Behnken design (BBD). The data were analyzed using various statistical parameters like: F-value, degree of freedom (DF), sum of squares (SS), coefficient of variation (CV) and regression coefficient (R2). These parameters generate statistical data, which were further used for analysis of BBD/quadric models showing the significant value for the optimization of biodegradation. The resultant data were used to plot the response surfaces curves (Wang et al. 2017).
Identification and molecular characterization of isolate S1
The primary characterization of isolate S1 was done by Gram’s straining. The KB013 kit (HiMedia, Mumbai India) was used for biochemical identification of isolate. The genomic DNA of isolate S1 was isolated by alkaline lysis method (Wilson 2001). The qualitative analysis of DNA was done by using 1.0% agarose gel through electrophoresis (BioRad USA). The amplification of isolate’s genomic DNA was done by polymerase chain reaction (PCR) method using universal 16S rRNA primers (27F and 1492R), respectively. After that, the amplified PCR product was sequenced (Eurofins Genomics India Pvt Ltd.). The sequence obtained after 16S rRNA was analyzed for presence of any chimeric sequence by DECIPHER version 1.12.2, an online bioinformatics tool. The Nucleotides Basic Local Alignment Search Tool (BLASTn) was further used for comparing isolate S1 sequence to the GenBank database of National Centre of Biotechnology Information (NCBI). Similar sequences were aligned with MUSCLE. The neighbor-joining (NJ) method was used to construct the phylogenetic tree using MEGA 7.0 software. The UGPMA clustering method was used for analyses of genetic variances between the sequences (Kumar et al. 2016b). The evolutionary history of isolate S1 was inferred using the neighbor-joining method (Saitou and Nei 1987).
Biodegradation study in pilot reactor
Batch experiment and kinetic study
The colorimetric method was used for detection of degradation percentage of APAP by the isolate S1. The 500 µL of degraded sample was mixed with 1.0 mL of 15% trichloroacetic acid (TCA) and centrifuged until a clear supernatant was formed. After this, the supernatant was transferred to another tube containing 0.5 mL of 6 N HCl and sodium nitrite (0.4 mL). This reaction produced nitrous acid which was neutralized by adding 15% sulfamic acid and finally add 15% sodium hydroxide (NaOH). The UV-spectrophotometer was used to detect absorbance of final mixture at 254 nm against water as blank (Shervington and Sakhnini 2000; Shihana et al. 2010). The degradation percentage (R) of APAP was calculated by Eq. 1:
Here \(C_{0}\) is absorbance at the initial concentration of APAP; Ct is the absorbance after incubation at time ‘t’.
Kinetic studies were performed by using same anaerobic reaction conditions mentioned above for isolate S1 with 100 to 2200 mg/L of APAP added in stepwise manner. The mixture was pipetted out after regular time interval of 4 h. Then cell growth in mixture was measured by using UV-spectrophotometer at optical density (OD600). The biomass dry weight was determined by filtering the cell suspension through a 0.2-μm-pore filter and then drying the filter to a constant weight for 24 h at 80 °C. Time course analysis of APAP biodegradation (APAP concentration vs. reaction time) was plotted. The experimental results were further used in various models but Haldane’s growth kinetics model for its best fit. This growth model was modified version of the Monod kinetics model. The kinetic analysis of the growth data was fitted in Eq. 2:
Here, ‘μ’ is the specific growth rate of strain S1; μmax is maximum specific growth rate of strain after time ‘t’; ‘S’ is the concentration of APAP (mg/L); ‘Ks’ is half-saturation constant (mg/L) and ‘Ki’ is inhibition constant (mg/L). To determine the yield coefficient by the strain S1, linear regression was used by accessing Eq. 3:
where ‘X’ and ‘X0’ are the biomass of strain S1 at time ‘t’ and initial concentration of biomass strain S1 (mg/L), respectively, and ‘S’ and ‘S0’ are the APAP concentration after time ‘t’ and initial concentration of APAP (mg/L), respectively.
The data analysis software Origin 2017 was used for analysis of experimental data on daily bases based on values of biomass concentration with time and fitted in the above equation. To perform the batch experiment waste water was collected from the Sonipat, Haryana (India). The effluent had unpleasant smell and was black in color. The batch experiment was performed with sterilized waste water supplemented with APAP (400 mg/L) in 2-L flask inoculated with isolate S1. No other nutrients were added in batch experiment. The degradation was observed after regular interval of every 4 h.
Biodegradation study in pilot reactor
The process of anaerobic biodegradation of APAP (400 mg/L) was also studied at pilot lab-scale 20-L fabricated reactor (Fig. 1e). The pilot unit was set with waste water collected from the sewage. The waste water was transferred to laboratory and used immediately. The COD, odor, color and pH were observed before reactor study and also during the reactor study as well. The UV-spectrophotometer-based method was used for COD detection. The reactor was fed with the sterilized waste water and distilled water (60:40). The absorbance was noted at UV 244 and supplemented with MSM (4 g/L) essential for microbial growth (Hesnawi et al. 2014; Joss et al. 2006). An overnight grown culture of S1 strain was inoculated in reactor. The samples were withdrawn after 5 days of interval of time and these observations were recorded for 30 days. The samples were analyzed for physiological changes in color of the medium during degradation, pH, COD analysis, biomass identification and biodegradation was also determined.
Biodegradation of APAP by bacterial strain S1 using batch culture study and morphology of the colony observed after 48 h on plate containing mineral salt medium (MSM) containing APAP. a The strain S1 inoculated with BHM and supplemented with 5 mg/mL of APAP; b after 48 h of incubation of the culture the appearance of the medium changes from colorless to light brown; c similarly after 5 days of incubation the color changed from light brown to black; d morphology of the colony observed after 48 h on plate containing mineral salt medium (MSM) containing APAP, e pilot-scale anaerobic reactor used for biodegradation of APAP
Identification of metabolites
The analysis of biodegrading products in the shake-flask study was performed by Fourier transform-infrared spectroscopy (FTIR) and gas chromatography–mass spectrometry (GC–MS) based techniques. This has led to identification of metabolic intermediates produced during degradation. FTIR spectroscopy has also been a top choice for minimizing the environmental issues regarding industrial chemical waste as it does not require much solvent. The structure of paracetamol contains different functional groups including –NH, O–H, C=O and aromatic ring containing C=C. The band appearing for C=O was selected for quantification as the interference arising from the excipients in pharmaceutical formulation is minimal in the region (Mallah et al. 2015). The 5 mL of degraded sample was filtered with Whatman paper (90 mm pore size) and centrifuged at 4000 rpm for 20 min. Then, the supernatant was transferred to a fresh tube containing equal volume of hexane by the gentle shaking of tube (Granberg and Rasmuson 1999). This tube was again centrifuged at 4000 rpm for 20 min. After second time centrifugation, the two separate layers appeared. The upper layer was taken in a fresh Eppendorf tube. This sample was stored at 4 °C and further used for FTIR and GC–MS analysis. FTIR was performed by Bruker instrument at 250–8000 cm−1.
The hexane mix metabolites were analyzed by GC–MS (Shimadzu-QP-2010 plus thermal desorption system T-20) at Advanced Instrumentation Research Facility (AIRF), JNU New Delhi, India. The GC–MS characterization of metabolites was performed with operation condition at 70 eV (electron impact mode); DDVP (Dichlorvos) column flow rate for single (1.7 mL/min) and double (3.4 mL/min) was used; helium as carrier gas (99.9%) was injected at 0.5 EL injection volume with 10:1 split ratio at injector temperature (250 °C); ion-source temperature (280 °C). The oven temperature was programmed to 110 °C for 2 min then increase in temperature with the rate of 10 °C/min to achieve 200 °C and further increase with rate of 5 °C/min till 280 °C was achieved and ends with at 280 °C for 9 min. The 70 eV mass spectra were taken with scanning interval of 0.5 s with fragments of 40 to 550 Da. PathPred, an online web-based tool was used to predict pathways from a query compounds as detected by GC–MS analysis of sample. The results interpretation through this server provides all the possible reactions in the form of tree indicated by the reaction pathways.
Results and discussion
Isolation and screening of APAP-degrading isolates
The conical flasks having control, test and abiotic sample were incubated at 37 °C in BOD incubator with shaking speed of 150 rpm. The color change was analyzed visually from white to brown and black was recorded after 2 and 5 days in flask containing active sewage sample (Fig. 1a–d). No physiological change was reported in control and test flasks. The inoculum was grown on plates containing MSM supplemented with APAP; the plates were then incubated at 37 °C in BOD incubator for 48 h. The color of media changed to black after 24 h and no individual colony was seen on the plate. A loop full of bacteria growing on plate was taken and cultured on plate containing MSM and APAP. After 48 h, individual bacterial colonies were observed and the medium around the colony turned brown/black (Fig. 1b, c). The physical appearances of all the colonies recovered were similar. The individual colony was re-cultured on nutrient agar plates (Fig. 1d). After incubation, a white color creamy colony was observed on the nutrient agar medium and this isolate was named as isolate S1. The isolate S1 does not require any other growth factors or supplements to grow in nutrient broth. It grows as monospecies on MSM supplemented with APAP as sole carbon, energy and nitrogen source. Many studies reported using microbial species that utilized xenobiotic compounds as a source of energy. Kumar et al. (2016a) reported the isolation and characterization of Kocuria sp. strain DAB-1 W, Staphylococcus sp. strain DAB-1Y and three Bacillus sp. strains uses gamma-HCH as carbon and energy source (Pannu and Kumar 2017). The adaptation of bacterial strain S1 with APAP, as carbon and nitrogen source occur in liquid media supplemented with APAP. The factors like concentration of APAP, temperature, pH, shaking speed (in rpm) played role in degradation determined through screening in shake-flask study (data unpublished). Further, these factors were optimized using the BBD and response surface methodology plots.
Experimental design, optimization studies and degradation analysis
The Design-Expert® software was used with the Box–Behnken design (BBD) with quadric model for predict the APAP degradation. The degrading efficiency of desired APAP was analyzed by putting the experimental value in the model and the predicted conditions of temperature, pH, agitation speed and APAP concentration were determined. The RSM was used to determine the optimization curve for the biodegradation of APAP. There was 29 values were predicted for four optimal condition. These 29 predicted values of experiments were performed in triplicate in conical flasks. The observed results for the predicted values of APAP degradation were analyzed by the software and their responses were predicted (Table 1). Further, these experimental results and predicted results were put into the model and RSM 3-D plots and contour curves were prepared. These 3-D plots show the effect of physical factor on degradation of APAP. The response surfaces values was fitted with Eq. (2).
Here, Y is predicted response (APAP degradation), A (pH), B (temperature), C (APAP concentration) and D (agitation speed) were the independent variables and they ultimately decides the fate of degradation. The predicted and experimental values of APAP concentration were analyzed with help of Design-Expert® (Table 1). The R2 was observed as response value defined by experimental factors and interactions between them as in Fig. 2a, showing a satisfactory correlation between residual and predicted values. The R2 for APAP degradation was 0.9510, which explains the variability of response up to 95.10%. The Adj R2 = 0.9019 was high for significance of the model. Predicted R2 of 0.7308 was realistic concurrence with Adj R2. The signal-to-noise ratio was measured by adequate precision the desirable ratio was greater and adequate precision ratio was of 14.355. The design space was navigated by model and the P and F values were determined by comparing of significance of each coefficient (Table 2). The ANOVA quadratic model demonstrates that this model was significant. The interaction of variable coefficients was not significant in determination of the response. The P value was less than 0.0001 and F-value was less than 0.05. This indicated that the models were significant. The P-value indicates highly significant models if value was less than 0.0522. The important model terms are B, D, AB, AC, AD and BD and in linear effect D (agitation speed) was one of the most important factors in optimization study, while C (APAP concentration) did not have much effect on the identified responses and Eq. 2 modified into Eq. (3).
Parity plot and 3D plots of RSM showing optimization study of degradation of APAP by strain S1 under different physiological conditions. a Parity plot showing distribution of experimental and predicted values of APAP; b 3D-RSM plot showing the effect of concentration of APAP vs. shake speed; c 3D RSM plot showing the effect of temperature vs. pH; d 3D- RSM plot showing the effect of shaking speed vs. pH
The function of two factors was determined by RSM plot. All the other factors were helpful at fixed level to understand the interactions among these factors. The residual vs predicted is shown in Fig. 2a. Response surface curves for APAP degradation between concentration of APAP vs speed (Fig. 2b), temperature vs pH (Fig. 2c), agitation speed vs pH (Fig. 2d), concentration vs temperature and agitation speed vs temperature. These results indicate the effect of four independent variables; temperature, pH, APAP concentration and shaking speed. The effect of APAP degradation rate depends on temperature (50–60 °C) and pH (8–9). Further, the effects of APAP concentration and agitation speed on APAP degradation were also determined. It was found that the lower APAP concentration and low agitation speed have no significant effect on degradation predicted through model. The temperature (40 °C), pH (7.0), APAP concentration (300 mg/L) and agitation speed (165 rpm) were reported for the maximum degradation of APAP by strain S1 in shake-flask study. The PPCPs mixture containing APAP, salicylic acid, carbamazepine, diethyltoluamide, and crotamiton were studied in batch experiments with soil. The experimental data were described with the help of pseudo-second-order kinetics having R2 > 0.98. The results indicated that there was effect of pH on adsorption of PPCPs mixture (Foolad et al. 2015).
Isolation, identification and molecular characterization of S1 strain
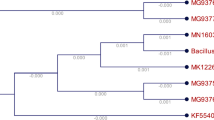
The isolate S1 was a facultative anaerobe Gram-positive bacteria having single and paired narrow tapered rods. These colonies were cream colored at the center and produce brownish pigment around the colony and were oxidase and catalase positive, non-spore forming, coccobacilli-shaped bacteria. Positive results were shown for malonate, citrate, catalase and arginine utilization. This strain shows variable biochemical result of Voges–Proskauer reaction and nitrate reduction, while ONPG, sucrose, mannitol, glucose, arabinose and trehalose biochemical test shows negative results. Genomic DNA was extracted from the 24-h grown culture of isolate S1. A single band of DNA having high-molecular weight was observed. Genomic DNA was isolated and 16S rDNA gene was amplified with PCR using universal (27F and 1492R) primers. The 1500-bp band was observed after evaluation of PCR product on agarose gel. The purification of the excised band from the gel was done to remove the contaminants. The 16S rDNA sequence generated by forward and reverse sequencing of PCR product was consensus. The 16S rRNA sequencing was used for molecular identification of genomic DNA extracted from isolate S1. The 16S rRNA was checked for chimeric sequence. The sequences data were used for BLAST analysis which suggested the genus Bacillus drentensis strain S1. The strain was named as Bacillus drentensis strain S1 based on 16S rRNA partial sequence and 1478-bp nucleotide sequence was submitted to NCBI GenBank with accession number KY623719. A phylogenetic tree of Bacillus drentensis strain S1 was performed MEGA 7.0 with BLAST hits based on similarity of sequences with maximum identity score were selected and aligned with Clustal-W. The hit table shows the related similarity of our strain with other strains (Additional file 1: S1). MEGA 7.0 was used to construct distance matrix and phylogenetic tree between the neighboring sequences which led to strain identification (Fig. 3). The identification of evolutionary history of the strain S1 was determined with the Kimura 2 parameter-based maximum likelihood method. The bootstrap agreement of tree provides 1000 replicates which were less than partitions reproduced of 50% bootstrap. Less than half bootstrap replicates were collapsed. Heuristic search for the initial tree were obtained by applying neighbor-join (NJ) method. The Bio-NJ algorithms were used for matrix to estimate pairwise distances using maximum composite likelihood (MCL) approach, further the topology was selected for better-quality log likelihood value. This analysis was performed with 11 nucleotide sequences having 1463 positions of the final dataset through MEGA 7.0 software. The tree constructed with the sum of branch length. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al. 2004) and are in the units of the number of base substitutions per site. This analysis involved 16 nucleotide sequences. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016b).
Heyrman et al. (2004) isolated Bacillus drentensis sp. (AJ542506) strain from an agricultural research area in Netherlands and reported it as Gram-positive anaerobic, tapered rods with cream-colored colonies producing brownish soluble pigment with butyrous consistency having eggshell-like structure. These features are similar to the strain Bacillus drentensis strain S1 (KY623719). This strain also produces brown pigment during the growth and degradation process. Kim et al. (2014) isolated Bacillus drentensis sp. strain from heavy metal-contaminated soils from a mine in Korea. The study on utilization of APAP as carbon and nitrogen sources was reported in Penicillium sp. by Hart and Orr (1975). Bart et al. (2011) reported Delftia tsuruhatensis and Pseudomonas aeruginosa that utilized APAP in membrane bioreactor (MBR), while Stenotrophomonas and Pseudomonas utilized paracetamol as energy sources (Zhang et al. 2013). The degradation of PPCPs and electricity generation were achieved by solid plain-graphite plate microbial fuel cell. The bacterial species Hydrogenophaga sp., Rubrivivax sp. and Leptothrix sp. were involved in PPCPs biodegradation (Bart et al. 2011).
Biodegradation study in batch and pilot reactor
Batch experiment and kinetic study
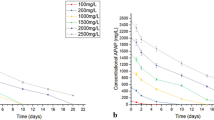
During the start of degradation, no lag phase was observed up to 458.4 ± 17.8 mg/L of APAP concentration. After 48 h, decrease was observed up to 50% concentration of APAP and the color change to brown was observed after 5 days and no APAP was detected (< 100 ng/L). No extra additives were added in effluent. Kinetic analysis was done in batch experiment using strain S1 and the biomass growth data required through variation in APAP concentration and semi-logarithmic graph was plotted using these degradation data (Fig. 4a). The graph was plotted between the specific growth rate (μ) and APAP concentration. The specific growth rate for APAP-degrading isolate S1 increases with increase in concentration of APAP from 100 to 400 mg/L and further increase in concentration the specific growth rate was decreased up to 2200 mg/L and the highest growth was observed at 400 mg/L. The μmax, Ks and Ki were 0.21/h, 155 with standard error of 5% mg/L, and 315 with standard error (SE) of 5% mg/L, respectively, while R2 of 0.9827 (Fig. 4a).
Based on initial concentrations, a linear plot was observed after short lag phase, this indicated that APAP acts as a substrate for the exponential growth of culture. The maximum growth rate of strain HJ1012 was observed at 315 mg/L with R2 of 0.946, respectively (Hu et al. 2013). Pseudomonas sp. strain ST-4 isolated from activated sludge samples has adapted concentrations 50 to 500 ppm (Khan et al. 2006); whereas, the Stenotrophomonas sp. f1, Pseudomonas sp. f2 and fg-2 were isolated from paracetamol-degrading aerobic aggregate with APAP concentration of 400, 2500 and 2000 mg/L, respectively (Zhang et al. 2013). The strains of P. putida, P. cepacia, and P. acidovorans were grown in MSM supplemented with acetamide or phenylacetamide (Betz and Clarke 1973). The kinetics experiments follow first-order kinetics. The half-life decreased from 1.6- to 11.7-fold on comparisons to initial values, and it was noted that APAP degraded to a greater extent (Martínez-Hernández et al. 2016). Other studies have indicated the degradation of APAP from the wide range and our strain has similar degradation behavior and degraded 300 g/L of acetaminophen (Table 3).
The bioremediation-based processes increases the degradation by APAP by microbial strains, and this has been an alternative for chemical techniques used in conventional treatment systems (Zur et al. 2018a, b). The six microbial strains were identified as Acinetobacter, Pseudomonas, Sphingomonas and Bacillus genus, which have the potential to degrade APAP. But the Pseudomonas moorei KB4 was able to use 50 mg/L of APAP as carbon source (Zur et al. 2018a, b). Pharmaceutical compounds (acetaminophen, caffeine, sulfamethoxazole, naproxen and carbamazepine) are also present in natural soil (loamy sand). The soil microbes have the potential to transform such compounds. The sorption and biodegradation kinetics models describe the degradation behavior. The serial batch-type reactor having soil and water ratio of 1:4 was supplemented with pharmaceutical with 100 μg/L concentration (Martínez-Hernández et al. 2016).
Biodegradation study in pilot reactor
An anaerobic reactor was constructed on the basis of information available on several factors such as concentration of waste water, volume of inoculum, agitation speed, physical conditions, etc. (Zwain et al. 2017). The main parameter was the load rate of APAP for degradation. The anaerobic reactor reaction rate was increased in initial stage due to presence of volatile fatty acids in waste water. Higher loading rate caused many problems, like acidification and lower the efficiency of degradation (COD removal) (Liang et al. 2016; Wang et al. 2017). The anaerobic reactor degradation occurs in three stages: (1) adjustment phase, (2) initiation of degradation, and (3) steady-state degradation.
The adjustment of system takes almost 20 h during which the COD (2700 mg/L) reduced slowly. The COD decreasing rate depends on the rate of degradation. The color of load and smell intensity gradually increased with rate of reaction. This shows that the S1 strain gradually degrades the APAP in the new environment. The increased pH was reported with respect to increase in degradation rate. It has been observed that after 3 days approximately 20% COD was decreased (Fig. 4b). This indicated the start of degradation, and the biomass of S1 was increased with reference to increase in time. The COD was decreased manyfold as the degradation proceeds for more time. The rate of decrease of COD was approximate 80% within 20 days. The pilot reactor was stable and self-buffered during APAP degradation. Further, the pH of the reactor changes and the nutrients are consumed in the reactor due to microbial growth and development; this anaerobic reactor degradation is slow as compared to batch reactor. The rate of degradation was increased gradually with respect to adjustment stage. The color of load turned slight blackish and the odor of the load was becoming unpleasant. These conditions assured that the microbes were slowly adapting during the adjustment stage and now the degradation of APAP was started by the S1 strain. During steady stage there was slight decrease in COD reduction wrt time. This was due to presence of low APAP concentration in the system. Almost 94.5% of APAP was degraded by the strain S1 in anaerobic conditions. The COD decreasing to manyfolds demonstrated that the S1 strain was working effectively under stress conditions. The color turned black and odor of the degraded products was unbearable. Such physical changes reflect that there was some kind of chemical reactions occurring in the strain S1 in the presence of APAP. These chemical reactions make a path to the degradation of APAP. During the whole process going within the anaerobic reactor, there was a slight fluctuation reported in pH (6.4 to 7.2) (Fig. 4b). The consortium of microalgae–bacteria has the potential to degrade ketoprofen, APAP and aspirin mixture in photobioreactors. These microbes degrade approximately 95% analgesics mixture and reduce COD (Ivshina et al. 2006). The microbial fuel cell having solid plain-graphite plates was used for biodegradation of APAP, ibuprofen, and sulfamethoxazole. The PPCP-containing sewage was used for microbial degradation during which electricity generated. The COD and nitrogen removal efficiencies were achieved almost 97.20% and 83.75%, during degradation of mixture. The removal efficiency was almost 98.21% to 99.89%. The microbial groups involved in degradation were Dechloromonas sp., Sphingomonas sp., and Pseudomonas aeruginosa (Chang et al. 2014).
Identification of metabolites
Degradation mediated by the Bacillus drentensis strain S1 was analyzed by FTIR. The alkane section was evidenced by sp2 C–H stretch band at 3098.98 cm−1. The presence of aromatic rings was evidenced by several bands in the spectrum. The alkane-like carbon monoxides have a band at 2070.97 cm−1. The characteristic C=C vibration stretch were observed at 1621.56 cm−1. The overtone combination bands appear between 3681 and 4000 cm−1. The vibrations were detected between 4000 and 2500 cm−1 region due to hydroxide (O–H), carbon–hydrogen (C–H) and nitrogen–hydrogen (N–H) stretching. The stretching produced by hydroxide has a broad range of 3700–3600 cm−1, while NH stretching is between 3400 and 3300 cm−1. R–OH stretch between 2500 and 3000 cm−1. An out-of-plane C–H bending at 736.23 cm−1 was characteristic of para-substituted aromatic compounds and at 3550–3500 cm−1 for phenol O–H stretching (Fig. 5a, b). Studies indicated that FTIR was used to determine the bonding pattern in organic solvents and biological macromolecules. Mallah et al. (2015) analyzed that acetaminophen contains functional groups like N–H, C=O, O–H and aromatic ring. Burgina et al. (2004) has calculated the potential and kinetic energies of APAP molecules (–CH3, –C=O, –NH, –C6H4, –C–O, and –O–H) and hydrogen bonds (–O–H…O and –N–H…O) in the experimental spectra. They indicated OH, NH and CH bonds between the spectra of 2800–3600 cm−1. The variation in bond suggested that the Bacillus drentensis strain S1 degraded acetaminophen.
Biodegradation of APAP gives many intermediate metabolites which were analyzed in a hexane extract by GC–MS. The gas chromatogram showed 66 peaks and the each peak indicates a single metabolite eluting close together (retention times of 17.709 to 51.510). Mass spectroscopy generates a library in the form of gas chromatogram peaks (Fig. 6a). The results of GC–MS indicated that, there was no residue of APAP existed in the batch culture. This indicated that Bacillus drentensis strain S1 utilizes APAP as energy source and has ability to remove APAP from the waste water. The metabolites of the APAP catabolic pathway were conclusively identified as oxalic acid, 2-isopropyl-5-methylcyclohexanone, and phenothiazine based on peak and retention time (RT) of sample (Fig. 6b, c).
After batch incubation study, the intermediates were obtained during degradation of APAP by the strain S1 analyzed by GC–MS. The derivatives obtained by mass spectra library were confirmed by published reports on APAP. The degradation of APAP analyzed by GC–MS led to the identification of compounds as oxalic acid and 2-isopropyl-5-methylcyclohexanone. These were identified on the basis of query run on PathPred server (Additional file 2: S2). This server predicts pathways from a query compounds (Additional file 3: S3). Oxalic acid and 2-isopropyl-5-methylcyclohexanone were detected as intermediates and GC–MS chromatogram in degrading sample mediated by S1 strain (Fig. 7). The retention time for oxalic acid and 2-isopropyl-5-methylcyclohexanone was 45.372 and 45.003, respectively (Table 4). Moreover, the Bacillus drentensis strain S1 shows mineralization of nitrogen of APAP to nitrites and nitrates. These compounds may also be indicators for APAP degradation and mineralization.
The metabolic pathway for APAP degradation by microbes was proposed via formation of 4-aminophenol to hydroquinone, which was considered as the major degradation route for APAP biodegradation. In hydroquinone pathway APAP was converted into 1,4-benzenediol or hydroquinone. The amino group of APAP was replaced by hydroxyl group during catalysis by Amidohydrolase which formulates hydroquinone. The hydroquinone subsequently transformed into fission rings while hydrolytic enzymes potentially catalyzed the hydroxylation of APAP during degradation, and potentially released acetamide to form hydroquinone which was further converted into 2-isopropyl-5-methylcyclohexanone and oxalic acid. The hydroquinone pathway and hydrolytic enzyme pathway for APAP degradation was purposed by Zhang et al. (2013). Other possible pathway for APAP degradation the hydroxylase oxidized APAP to form N-(4-hydroxyphenyl)-acetamide by the replacement of hydrogen to produce p-aminophenol. The Burkholderia sp. strain AK-5 follows the hydroquinone and pyrocatechol degradation pathway (Takenaka et al. 2003). APAP form dimer and trimer during elimination pathway in dark incubation (Liang et al. 2016) intermediate products p-aminophenol and hydroquinone were recognized by LC/MS. Physical factors like pH, temperature do not have a prominent effect on the degradation mediated by KB4 strain (Zur et al. 2018a, b).
Conclusion
Acetaminophen is a common drug used extensively as a pain reliever and a fever reducer. Due to its regular use its concentration gets increased in our environment gradually. Therefore, there is an urgent need to eliminate such contaminants from the environment. Biodegradation is one of the best approaches to remediate such contaminants. Bacillus drentensis strain S1 is an isolate identified from sewage sample having the potential to remove acetaminophen from waste water. This strain also degrades and tolerates the wide range of APAP concentration. The degrading potential of strain in anaerobic pilot-scale reactor was slow compared to the batch reactor. It is due to the fluctuation in pH and exhaustion of nutrients. We cannot underestimate the potential of strain, because it is an eco-friendly and cost-effective approach for degradation of APAP. It also has ability to degrade the APAP as sole carbon and energy source. Acetaminophen degradation pathways gave us indication of kind of intermediates produced during biodegradation, but sometimes biotransformation/biodegradation leads to production of dead-end metabolites. Therefore, the long-term prospective for the acetaminophen removal from the environment is needed and Bacillus drentensis strain S1 can play the major role. The Bacillus drentensis strain S1 strain has the potential to lead in acetaminophen removal from various water sources. With the aid of modern technology using nanotechnology, biosensors, and deploying better degrading strains, etc., can increase the degradation potential of harmful compounds in the environment.
Availability of data and materials
The datasets supporting this article are included in the manuscript.
Abbreviations
- PPCPs:
-
Pharmaceuticals and personal care products
- HCl:
-
Hydrochloric acid
- GC–MS:
-
Gas chromatography–mass spectroscopy
- RSM:
-
Response surface methodology
- FT-IR:
-
Fourier transform-infrared spectroscopy
- BBD:
-
Box–Behnken design
- NSAIDs:
-
Nonsteroidal anti-inflammatory drugs
- EOCs:
-
Emerging organic contaminants
- WTPs:
-
Waste water treatment plants
- RT:
-
Retention time
References
Balakrishna K, Rath A, Praveenkumarreddy Y, Guruge KS, Subedi B (2017) A review of the occurrence of pharmaceuticals and personal care products in Indian water bodies. Ecotoxicol Environ Saf 137:113–120. https://doi.org/10.1016/J.ECOENV.2016.11.014
Bart DG, Vanhaecke L, Verstraete W, Boon N (2011) Degradation of acetaminophen by Delftia tsuruhatensis and Pseudomonas aeruginosa in a membrane bioreactor. Water Res 45(4):1829–1837. https://doi.org/10.1016/J.WATRES.2010.11.040
Bedner M, MacCrehan WA (2006) Transformation of acetaminophen by chlorination produces the toxicants 1,4-benzoquinone and N-acetyl-p-benzoquinone imine. Environ Sci Technol 40(2):516–522. https://doi.org/10.1021/es0509073
Betz JL, Clarke PH (1973) Growth of pseudomonas species on phenylacetamide. J Gen Microbiol 75:167–177. https://doi.org/10.1099/00221287-75-1-167
Brillas E, Sirés I, Arias C, Cabot PL, Centellas F, Rodríguez RM, Garrido JA (2005) Mineralization of paracetamol in aqueous medium by anodic oxidation with a boron-doped diamond electrode. Chemosphere 58(4):399–406. https://doi.org/10.1016/J.CHEMOSPHERE.2004.09.028
Burgina EB, Baltakhinov VP, Boldyreva EV, Shakhtschneider TP (2004) IR spectra of paracetamol and phenacetin. 1. Theoretical and experimental studies. J Struct Chem 45:64–73. https://doi.org/10.1023/B:JORY.0000041502.85584.d5
Chandrasekharan NV, Dai H, Roos KLT, Evanson NK, Tomsik J, Elton TS, Simmons DL (2002) COX-3, a cyclooxygenase-1 variant inhibited by acetaminophen and other analgesic/antipyretic drugs: cloning, structure, and expression. Proc Natl Acad Sci 99(21):13926–13931. https://doi.org/10.1073/pnas.162468699
Chang YT, Yang CW, Chang YJ, Chang TC, Wei DJ (2014) The treatment of PPCP-containing sewage in an anoxic/aerobic reactor coupled with a novel design of solid plain graphite-plates microbial fuel cell. Biomed Res Int 2014:765652. https://doi.org/10.1155/2014/765652
Chen CY, Chen SC, Fingas M, Kao CM (2010) Biodegradation of propionitrile by Klebsiella oxytoca immobilized in alginate and cellulose triacetate gel. J Hazard Mater 177:856—863. https://doi.org/10.1016/j.jhazmat.2009.12.112
Edrees WH, AL-Kaf AG, Abdullah QY, Naji KM (2018) Isolation and identification of a new bacterial strains degrading Paracetamol isolated from Yemeni Environment. Clin Biotechnol Microbiol 1(6):257–270. https://www.scientiaricerca.com/srcbmi/SRCBMI-01-00039.php%0A https://www.scientiaricerca.com/srcbmi/pdf/SRCBMI-01-00039.pdf
Foolad M, Hu J, Tran NH, Ong SL (2015) Sorption and biodegradation characteristics of the selected pharmaceuticals and personal care products onto tropical soil. Water Sci Technol 73(1):51–59. https://doi.org/10.2166/wst.2015.461
Granberg RA, Rasmuson ÅC (1999) Solubility of paracetamol in pure solvents. J Chem Eng Data 44(6):1391–1395. https://doi.org/10.1021/je990124v
Hart A, Orr DLJ (1975) The degradation of paracetamol (4-hydroxyacetanilide) and other substituted acetanilides by a Penicillium species. Antonie Van Leeuwenhoek 41(1):239–247. https://doi.org/10.1007/BF02565059
Hasan Z, Jeon J, Jhung SH (2012) Adsorptive removal of naproxen and clofibric acid from water using metal-organic frameworks. J Hazard Mater 209–210:151–157. https://doi.org/10.1016/j.jhazmat.2012.01.005
Hesnawi R, Dahmani K, Al-Swayah A, Mohamed S, Mohammed SA (2014) Biodegradation of municipal waste water with local and commercial bacteria. Procedia Eng 70:810–814. https://doi.org/10.1016/J.PROENG.2014.02.088
Heyrman J, Vanparys B, Logan NA, Balcaen A, Rodríguez-Díaz M, Felske A, De Vos P (2004) Bacillus novalis sp. nov., Bacillus vireti sp. nov., Bacillus soli sp. nov., Bacillus bataviensis sp. nov. and Bacillus drentensis sp. nov., from the Drentse A grasslands. Int J Syst Evol Microbiol 54(1):47–57. https://doi.org/10.1099/ijs.0.02723-0
Hu J, Zhang LL, Chen JM, Liu Y (2013) Degradation of paracetamol by Pseudomonas aeruginosa strain HJ1012. J Environ Sci Health Part A 48(7):791–799. https://doi.org/10.1080/10934529.2013.744650
Ismail MM, Essam TM, Ragab YM, El-Sayed AE-KB, Mourad FE (2017) Remediation of a mixture of analgesics in a stirred-tank photobioreactor using microalgal-bacterial consortium coupled with attempt to valorise the harvested biomass. Bioresour tTechnol 232:364–371. https://doi.org/10.1016/j.biortech.2017.02.062
Ivshina IB, Rychkova MI, Vikhareva EV, Chekryshkina LA, Mishenina II (2006) Catalysis of the biodegradation of unusable medicines by Alkanotrophic rhodococci. Appl Biochem Microbiol 42(4):392–395. https://doi.org/10.1134/S0003683806040090
Jameson JE (1961) A study of tetrathionate enrichment techniques, with particular reference to two new tetrathionate modifications used in isolating salmonellae from sewer swabs. J Hyg 59(1):1–13. https://doi.org/10.1017/S0022172400038663
Joss A, Zabczynski S, Göbel A, Hoffmann B, Löffler D, McArdell CS et al (2006) Biological degradation of pharmaceuticals in municipal waste water treatment: proposing a classification scheme. Water Res 40(8):1686–1696. https://doi.org/10.1016/J.WATRES.2006.02.014
Khan AS, Hamayun M, Ahmed S (2006) Degradation of 4-aminophenol by newly isolated Pseudomonas sp. strain ST-4. Enzyme Microb Technol 38(1–2):10–13. https://doi.org/10.1016/j.enzmictec.2004.08.045
Khetan SK, Collins TJ (2007) Human pharmaceuticals in the aquatic environment: a challenge to green chemisty. Chem Rev 107(6):2319–2364. https://doi.org/10.1021/cr020441w
Kim I, Lee M, Wang S (2014) Heavy metal removal in groundwater originating from acid mine drainage using dead Bacillus drentensis sp. immobilized in polysulfone polymer. J Environ Manage 146:568–574. https://doi.org/10.1016/J.JENVMAN.2014.05.042
Kumar D, Kumar A, Sharma J (2016a) Degradation study of lindane by novel strains Kocuria sp. DAB-1Y and Staphylococcus sp. DAB-1W. Bioresour Bioprocess. https://doi.org/10.1186/s40643-016-0130-8
Kumar S, Stecher G, Tamura K (2016b) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Li J, Ye Q, Gan J (2014) Degradation and transformation products of acetaminophen in soil. Water Res 49:44–52. https://doi.org/10.1016/J.WATRES.2013.11.008
Liang C, Lan Z, Zhang X, Liu Y (2016) Mechanism for the primary transformation of acetaminophen in a soil/water system. Water Res 98:215–224. https://doi.org/10.1016/J.WATRES.2016.04.027
Mallah MA, Sherazi STH, Bhanger MI, Mahesar SA, Bajeer MA (2015) A rapid Fourier-transform infrared (FTIR) spectroscopic method for direct quantification of paracetamol content in solid pharmaceutical formulations. Spectrochim Acta Part A Mol Biomol Spectrosc 141:64–70. https://doi.org/10.1016/j.saa.2015.01.036
Martínez-Hernández V, Meffe R, Herrera López S, de Bustamante I (2016) The role of sorption and biodegradation in the removal of acetaminophen, carbamazepine, caffeine, naproxen and sulfamethoxazole during soil contact: a kinetics study. Sci Total Environ 559:232–241. https://doi.org/10.1016/J.SCITOTENV.2016.03.131
Minto CF, Schnider TW, Egan TD, Youngs E, Lemmens HJ, Gambus PL et al (1997) Influence of age and gender on the pharmacokinetics and pharmacodynamics of remifentanil: I. Model development. Anesthesiology 86(1):10–23
Mirizadeh S, Yaghmaei S, Ghobadi Nejad Z (2014) Biodegradation of cyanide by a new isolated strain under alkaline conditions and optimization by response surface methodology (RSM). J Environ Health Sci Eng 12(1):85. https://doi.org/10.1186/2052-336X-12-85
Mutnur S (2014) Bioremediation of paracetamol from industrial waste water by Pseudomonas mendocina. In: 12th Specialized conference on small water and waste water systems and 4th specialized conference on resources oriented sanitation, November 2–4, 2014 Muscat, Sultanate of Oman
Onesios KM, Yu JT, Bouwer EJ (2009) Biodegradation and removal of pharmaceuticals and personal care products in treatment systems: a review. Biodegradation 20(4):441–466. https://doi.org/10.1007/s10532-008-9237-8
Pannu R, Kumar D (2017) Process optimization of γ-hexachlorocyclohexane degradation using three novel Bacillus sp. strains. Biocatal Agric Biotechnol 11:97–107. https://doi.org/10.1016/J.BCAB.2017.06.009
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shervington LA, Sakhnini N (2000) A quantitative and qualitative high performance liquid chromatographic determination of acetaminophen and five of its para-substituted derivatives. J Pharm Biomed Anal 24(1):43–49. https://doi.org/10.1016/S0731-7085(00)00396-4
Shihana F, Dissanayake D, Dargan P, Dawson A (2010) A modified low-cost colorimetric method for paracetamol (acetaminophen) measurement in plasma. Clin Toxicol 48(1):42–46. https://doi.org/10.3109/15563650903443137
Skoumal M, Cabot P-L, Centellas F, Arias C, Rodríguez RM, Garrido JA, Brillas E (2006) Mineralization of paracetamol by ozonation catalyzed with Fe2+ , Cu2+ and UVA light. Appl Catal B 66(3–4):228–240. https://doi.org/10.1016/J.APCATB.2006.03.016
Subedi B, Balakrishna K, Joshua DI, Kannan K (2017) Mass loading and removal of pharmaceuticals and personal care products including psychoactives, antihypertensives, and antibiotics in two sewage treatment plants in southern India. Chemosphere 167:429–437. https://doi.org/10.1016/J.CHEMOSPHERE.2016.10.026
Takenaka S, Murakami S, Shinke R, Aoki K (1998) Metabolism of 2-aminophenol by Pseudomonas sp. AP-3: modified meta-cleavage pathway. Arch Microbiol 170(2):132–137. https://doi.org/10.1007/s002030050624
Takenaka S, Okugawa S, Kadowaki M, Murakami S, Aoki K (2003) The metabolic pathway of 4-aminophenol in Burkholderia sp strain AK-5 differs from that of aniline and aniline with C-4 substituents. Appl Environ Microbiol 69(9):5410–5413. https://doi.org/10.1128/aem.69.9.5410-5413.2003
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035
Vogna D, Marotta R, Napolitano A, Ischia M (2002) Advanced oxidation chemistry of paracetamol. UV/H2O2-induced hydroxylation/degradation pathways and 15N-aided inventory of nitrogenous breakdown products. J Org Chem 67(17):6143–6151. https://doi.org/10.1021/jo025604v
Wang Y, Wang Q, Wang Y, Han H, Hou Y, Shi Y (2017) Statistical optimization for the production of recombinant cold-adapted superoxide dismutase in E. coli using response surface methodology. Bioengineered 8(6):693–699. https://doi.org/10.1080/21655979.2017.1303589
Waterston K, Wang JW, Bejan D, Bunce NJ (2006) Electrochemical waste water treatment: electrooxidation of acetaminophen. J Appl Electrochem 36(2):227–232. https://doi.org/10.1007/s10800-005-9049-z
Wei F, Zhou Q-W, Leng SQ (2011) Isolation, identification and biodegradation characteristics of a new bacterial strain degrading paracetamol. Chin J Environ Sci 32(6):1812–1819
Wilson K (2001) Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol 56(1):2.4.1–2.4.5. https://doi.org/10.1002/0471142727.mb0204s56
Wu S, Zhang L, Chen J (2012) Paracetamol in the environment and its degradation by microorganisms. Appl Microbiol Biotechnol 96(4):875–884. https://doi.org/10.1007/s00253-012-4414-4
Yang L, Yu LE, Ray MB (2008) Degradation of paracetamol in aqueous solutions by TiO2 photocatalysis. Water Res 42(13):3480–3488. https://doi.org/10.1016/J.WATRES.2008.04.023
Yang D, Liu H, Zheng Z, Yuan Y, Zhao JC, Waclawik ER et al (2009) An efficient photocatalyst structure: TiO2(B) nanofibers with a shell of anatase nanocrystals. J Am Chem Soc 131(49):17885–17893. https://doi.org/10.1021/ja906774k
Zhang L, Hu J, Zhu R, Zhou Q, Chen J (2013) Degradation of paracetamol by pure bacterial cultures and their microbial consortium. Appl Microbiol Biotechnol 97(8):3687–3698. https://doi.org/10.1007/s00253-012-4170-5
Żur J, Piński A, Marchlewicz A, Hupert-Kocurek K, Wojcieszyńska D, Guzik U (2018a) Organic micropollutants paracetamol and ibuprofen—toxicity, biodegradation, and genetic background of their utilization by bacteria. Environ Sci Pollut Res 25(22):21498–21524. https://doi.org/10.1007/s11356-018-2517-x
Żur J, Wojcieszyńska D, Hupert-Kocurek K, Marchlewicz A, Guzik U (2018b) Paracetamol toxicity and microbial utilization. Pseudomonas moorei KB4 as a case study for exploring degradation pathway. Chemosphere 206:192–202. https://doi.org/10.1016/J.CHEMOSPHERE.2018.04.179
Zwain HM, Aziz HA, Ng WJ, Dahlan I (2017) Performance and microbial community analysis in a modified anaerobic inclining-baffled reactor treating recycled paper mill effluent. Environ Sci Pollut Res 24(14):13012–13024. https://doi.org/10.1007/s11356-017-8804-0
Acknowledgements
The authors acknowledge the sample analysis for FTIR at Central Instrumentation Laboratory (CIL), DCRUST Murthal Sonepat India, DNA sequencing at Eurofins Genomics India Pvt Ltd, Advanced Instrumentation Research Facility (AIRF), JNU New Delhi, India, for GC–MS analysis. The author S. Chopra wish to thank UGC New Delhi, India, for providing research assistantship in the form of RGNF fellowship.
Funding
There is no external funding received to carry out this research. Authors wish to thank the Department of Biotechnology, DCRUST Murthal Sonipat India, for providing the necessary facilities to carry out this research.
Author information
Authors and Affiliations
Contributions
SC conducted the experiment and DK helped in the experiments and supervised the research. Both authors prepared the manuscript. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
Both authors approved this manuscript.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: S1.
Hit table of phylogenetic analysis of by Bacillus drentensis strain S1.
Additional file 2: S2.
Predicted pathway of degradation of APAP by Bacillus drentensis strain S1 predicted by the PathPred online tool with KEGG compound number.
Additional file 3: S3.
Different intermediates predicted through PathPred online tool with structure of predicted compounds.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chopra, S., Kumar, D. Characterization, optimization and kinetics study of acetaminophen degradation by Bacillus drentensis strain S1 and waste water degradation analysis. Bioresour. Bioprocess. 7, 9 (2020). https://doi.org/10.1186/s40643-020-0297-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40643-020-0297-x