Abstract
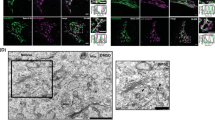
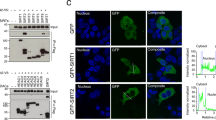
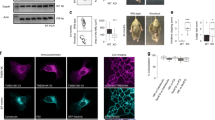
The retrograde transport inhibitor Retro-2 has a protective effect on cells and in mice against Shiga-like toxins and ricin. Retro-2 causes toxin accumulation in early endosomes and relocalization of the Golgi SNARE protein syntaxin-5 to the endoplasmic reticulum. The molecular mechanisms by which this is achieved remain unknown. Here, we show that Retro-2 targets the endoplasmic reticulum exit site component Sec16A, affecting anterograde transport of syntaxin-5 from the endoplasmic reticulum to the Golgi. The formation of canonical SNARE complexes involving syntaxin-5 is not affected in Retro-2-treated cells. By contrast, the interaction of syntaxin-5 with a newly discovered binding partner, the retrograde trafficking chaperone GPP130, is abolished, and we show that GPP130 must indeed bind to syntaxin-5 to drive Shiga toxin transport from the endosomes to the Golgi. We therefore identify Sec16A as a druggable target and provide evidence for a non-SNARE function for syntaxin-5 in interaction with GPP130.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data and code availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier project accession no. PXD015642. Imaris and Matlab scripts for EEA1 quantification are available on request. All other data supporting the findings of this study are available within the paper and its supplementary information files.
References
Johannes, L. & Römer, W. Shiga toxins—from cell biology to biomedical applications. Nat. Rev. Microbiol. 8, 105–116 (2010).
Tarr, P. I., Gordon, C. A. & Chandler, W. L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365, 1073–1086 (2005).
Endo, Y. et al. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 171, 45–50 (1988).
Fraser, M. E., Chernaia, M. M., Kozlov, Y. V. & James, M. N. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 Å resolution. Nat. Struct. Biol. 1, 59–64 (1994).
Ling, H. et al. Structure of Shiga-like toxin I B-pentamer complexed with an analogue of its receptor Gb3. Biochemistry 37, 1777–1788 (1998).
Sandvig, K., Olsnes, S., Brown, J. E., Petersen, O. W. & van Deurs, B. Endocytosis from coated pits of Shiga toxin: a glycolipid-binding protein from Shigella dysenteriae 1. J. Cell Biol. 108, 1331–1343 (1989).
Römer, W. et al. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 450, 670–675 (2007).
Mallard, F. et al. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156, 653–664 (2002).
Sandvig, K. et al. Retrograde transport of endocytosed Shiga toxin to the endoplasmic reticulum. Nature 358, 510–512 (1992).
Spooner, R. A. & Lord, J. M. How ricin and Shiga toxin reach the cytosol of target cells: retrotranslocation from the endoplasmic reticulum. Curr. Top. Microbiol. Immunol. 357, 19–40 (2012).
Johannes, L. & Popoff, V. Tracing the retrograde route in protein trafficking. Cell 135, 1175–1187 (2008).
Sandvig, K., Skotland, T., van Deurs, B. & Klokk, T. I. Retrograde transport of protein toxins through the Golgi apparatus. Histochem. Cell Biol. 140, 317–326 (2013).
Bujny, M. V., Popoff, V., Johannes, L. & Cullen, P. J. The retromer component, sorting nexin-1, is required for efficient early endosome-to-trans Golgi network retrograde transport of Shiga toxin. J. Cell Sci. 120, 2010–2021 (2007).
Popoff, V. et al. The retromer complex and clathrin define a post-early endosomal retrograde exit site. J. Cell Sci. 120, 2022–2031 (2007).
Utskarpen, A., Slagsvold, H. H., Dyve, A. B., Skanland, S. S. & Sandvig, K. SNX1 and SNX2 mediate retrograde transport of Shiga toxin. Biochem. Biophys. Res. Commun. 358, 566–570 (2007).
Mukhopadhyay, S. & Linstedt, A. D. Manganese blocks intracellular trafficking of Shiga toxin and protects against Shiga toxicosis. Science 335, 332–335 (2012).
Mukhopadhyay, S., Redler, B. & Linstedt, A. D. Shiga toxin binding site for host cell receptor GPP130 reveals unexpected divergence in toxin trafficking mechanisms. Mol. Biol. Cell 24, 2311–2318 (2013).
Tai, G. et al. Participation of syntaxin 5/Ykt6/GS28/GS15 SNARE complex in transport from the early/recycling endosome to the TGN. Mol. Biol. Cell 15, 4011–4022 (2004).
Hui, N. et al. An isoform of the Golgi t-SNARE, syntaxin 5, with an endoplasmic reticulum retrieval signal. Mol. Biol. Cell 8, 1777–1787 (1997).
Mancias, J. D. & Goldberg, J. Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. Embo J. 27, 2918–2928 (2008).
Gillet, D., Barbier, J., Johannes, L., Cintrat, J. -C. & Noël, R. New compounds having a protective activity against toxins with intracellular activity. Patent WO2014060586A1 (2012).
Mukhopadhyay, S. & Linstedt, A. D. Retrograde trafficking of AB(5) toxins: mechanisms to therapeutics. J. Mol. Med. (Berl.) 91, 1131–1141 (2013).
Stechmann, B. et al. Inhibition of retrograde transport protects mice from lethal ricin challenges. Cell 141, 231–242 (2010).
Secher, T. et al. Retrograde trafficking inhibitors of Shiga toxins reduces morbidity and mortality of mice infected with enterohemorrhagic Escherichia coli (STEC). Antimicrob. Agents Chemother. 59, 5010–5013 (2015).
Canton, J. & Kima, P. E. Targeting host syntaxin-5 preferentially blocks Leishmania parasitophorous vacuole development in infected cells and limits experimental Leishmania infections. Am. J. Pathol. 181, 1348–1355 (2012).
Gupta, N. et al. Inhibitors of retrograde trafficking active against ricin and Shiga toxins also protect cells from several viruses, Leishmania and Chlamydiales. Chem. Biol. Interact. 267, 96–103 (2017).
Carney, D. W. et al. Structural optimization of a retrograde trafficking inhibitor that protects cells from infections by human polyoma- and papillomaviruses. Bioorg. Med. Chem. 22, 4836–4847 (2014).
Dai, W. et al. Antiviral effects of Retro-2(cycl) and Retro-2.1 against Enterovirus 71 in vitro and in vivo. Antivir. Res. 144, 311–321 (2017).
Dai, W. W. Antiviral effect of Retro-2.1 against herpes simplex virus type 2. J. Microbiol. Biotechnol. 28, 849–859 (2018).
Cruz, L. et al. Potent inhibition of human cytomegalovirus by modulation of cellular SNARE syntaxin 5. J. Virol. 91, e01637-16 (2017).
Shtanko, O. et al. Retro-2 and its dihydroquinazolinone derivatives inhibit filovirus infection. Antivir. Res. 149, 154–163 (2018).
Harrison, K. et al. Vaccinia virus uses retromer-independent cellular retrograde transport pathways to facilitate the wrapping of intracellular mature virions during virus morphogenesis. J. Virol. 90, 10120–10132 (2016).
Sivan, G., Weisberg, A. S., Americo, J. L. & Moss, B. Retrograde transport from early endosomes to the trans-Golgi network enables membrane wrapping and egress of vaccinia virus virions. J. Virol. 90, 8891–8905 (2016).
Noel, R. et al. N-methyldihydroquinazolinone derivatives of Retro-2 with enhanced efficacy against Shiga toxin. J. Med. Chem. 56, 3404–3413 (2013).
Whittle, J. R. & Schwartz, T. U. Structure of the Sec13–Sec16 edge element, a template for assembly of the COPII vesicle coat. J. Cell Biol. 190, 347–361 (2010).
Wälchli, S. et al. The mitogen-activated protein kinase p38 links Shiga toxin-dependent signaling and trafficking. Mol. Biol. Cell. 19, 95–104 (2008).
Hughes, H. & Stephens, D. J. Assembly, organization, and function of the COPII coat. Histochem. Cell Biol. 129, 129–151 (2008).
Xu, Y., Martin, S., James, D. E. & Hong, W. GS15 forms a SNARE complex with syntaxin 5, GS28, and Ykt6 and is implicated in traffic in the early cisternae of the Golgi apparatus. Mol. Biol. Cell 13, 3493–3507 (2002).
Chen, Y. A. & Scheller, R. H. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2, 98–106 (2001).
Hong, W. SNAREs and traffic. Biochim. Biophys. Acta 1744, 493–517 (2005).
Amessou, M. et al. Syntaxin 16 and syntaxin 5 control retrograde transport of several exogenous and endogenous cargo proteins. J. Cell. Sci. 120, 1457–1468 (2007).
Puri, S., Bachert, C., Fimmel, C. J. & Linstedt, A. D. Cycling of early Golgi proteins via the cell surface and endosomes upon lumenal pH disruption. Traffic 3, 641–653 (2002).
Natarajan, R. & Linstedt, A. D. A cycling cis-Golgi protein mediates endosome-to-Golgi traffic. Mol. Biol. Cell 15, 4798–4806 (2004).
Linstedt, A. D., Mehta, A., Suhan, J., Reggio, H. & Hauri, H. P. Sequence and overexpression of GPP130/GIMPc: evidence for saturable pH-sensitive targeting of a type II early Golgi membrane protein. Mol. Biol. Cell 8, 1073–1087 (1997).
Ledger, P. W., Uchida, N. & Tanzer, M. L. Immunocytochemical localization of procollagen and fibronectin in human fibroblasts: effects of the monovalent ionophore, monensin. J. Cell Biol. 87, 663–671 (1980).
Mollenhauer, H. H., Morre, D. J. & Rowe, L. D. Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochim. Biophys. Acta 1031, 225–246 (1990).
Cañeque, T., Muller, S. & Rodriguez, R. Visualizing biologically active small molecules in cells using click chemistry. Nat. Rev. Chem 2, 202–215 (2018).
Adolf, F., Rhiel, M., Reckmann, I. & Wieland, F. T. Sec24C/D-isoform-specific sorting of the preassembled ER-Golgi Q-SNARE complex. Mol. Biol. Cell 27, 2697–2707 (2016).
Campbell, J. L. & Schekman, R. Selective packaging of cargo molecules into endoplasmic reticulum-derived COPII vesicles. Proc. Natl Acad. Sci. USA 94, 837–842 (1997).
Gupta, N. et al. (S)-N-methyldihydroquinazolinones are the active enantiomers of Retro-2 derived compounds against toxins. ACS Med. Chem. Lett. 5, 94–97 (2014).
Mallard, F. et al. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of Shiga toxin B-fragment transport. J. Cell Biol. 143, 973–990 (1998).
Jordan, M., Schallhorn, A. & Wurm, F. M. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24, 596–601 (1996).
Poullet, P., Carpentier, S. & Barillot, E. myProMS, a web server for management and validation of mass spectrometry-based proteomic data. Proteomics 7, 2553–2556 (2007).
Valot, B., Langella, O., Nano, E. & Zivy, M. MassChroQ: a versatile tool for mass spectrometry quantification. Proteomics 11, 3572–3577 (2011).
Vizcaino, J. A. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033 (2016).
Soderberg, O. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Boncompain, G. & Perez, F. Synchronizing protein transport in the secretory pathway. Curr. Protoc. Cell Biol. 57, 15.19.1–15.19.16 (2012).
Venditti, R. et al. Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science 337, 1668–1672 (2012).
Acknowledgements
We thank R. Rodriguez (cell imaging of Retro-2.1 and target identification), G. Boncompain (RUSH), C. Viaris De Lesegno (PLA), R. Onclercq-Delic and S. Bombard (intoxication assays), C. Brewee and B. Sancerne (recombinant protein production and cytotoxicity assays), D. Buisson (purification of 15) and E. Chirkin (synthesis of new batches of chemicals) for help with the indicated experiments. We thank V. Sabatet from the Laboratoire de Spectrométrie de Masse Protéomique for myProMS assistance. We acknowledge support from grants from the Agence Nationale pour la Recherche (ANR-11-BSV2-0018 and ANR-14-CE16-0004-03 to L.J., J.B., J.-C.C. and D.G. and ANR-19-CE13-0001-01 to L.J.), the Human Frontier Science Program (RGP0029-2014 to L.J.), the European Research Council (advanced grant no. 340485 to L.J.), the Swedish Research Council (K2015-99X-22877-01-6 to L.J., J.-C.C. and D.G.), the Joint Ministerial Program of R&D against CBRNE Risks (D.G., J.B., J.-C.C. and L.J.), the CEA (D.G., J.B. and J.-C.C.), the Île de France Region DIM Malinf initiative (grant no. 140101 to D.G., J.B. and L.J.), the Région Île-de-France (D.L.) and the Fondation pour la Recherche Médicale (D.L.). The Gillet and Cintrat teams are members of LabEx LERMIT (ANR-10-LABX-33) and the Johannes team is a member of Labex CelTisPhyBio (11-LBX-0038) and Idex Paris Sciences et Lettres (ANR-10-IDEX-0001–02 PSL). We also acknowledge the Cell and Tissue Imaging (PICT-IBiSA) and Nikon Imaging Centre, Institut Curie, a member of the French National Research Infrastructure France-BioImaging (ANR10-INBS-04).
Author information
Authors and Affiliations
Contributions
L.J. and D.G. conceived and designed the study. S.J.R., H.-F.R. and M.D.G.-C. performed the click chemistry immunofluorescence experiments and M.D.G.-C. and S.J.R. performed the click chemistry pulldown experiments. S.J.R. performed the in vitro Retro-2 pulldown, the SNARE PLA, the SNARE relocalization, the Syn5-RUSH assay and the GPP130 rescue analysis. S.J.R., M.D.G.-C., J.B. and L.T. performed the intoxication assays. The BLI assay was performed by J.B. and R.S. Purification of the Syn5 and GPP130 variants, the monensin study and the in vitro Syn5 pulldown of the GPP130 variants were performed by C.B. and A.D.L. S.J.R. and M.D.G.-C. performed the Sec16A and syntaxin-5 proteomics analysis and immunofluorescence. A.F. performed the Sec23 kinetic studies, the GFP-Sec16A pulldown and the STxB, GPP130 and Syn5 immunofluorescence with quantification. C.A.V.-C. wrote the scripts and automated the EEA1 colocalization methods. A.C., M.M. and J.-C.C. designed and performed the chemical synthesis of the azide-functionalized Retro-2 derivatives, and J.B. and L.T. characterized their anti-Shiga toxin activity. J.M., S.P. and L.T. prepared and characterized the recombinant Sec16A1266–1678/Sec13 protein complex. F.D. carried out the MS work, and D.L. supervised the MS and proteomic data analysis. S.J.R. and L.J. wrote the paper. A.F., J.-C.C., J.B., D.G., A.D.L. and C.L. critically revised the manuscript and aided in the design and analysis of experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–2, Figs. 1–7 and Supplementary Note.
Supplementary Dataset 1
Proteomics quantification results GFP-Sec16A vehicle versus EGFP.
Supplementary Dataset 2
Proteomics quantification results GFP-Sec16A Retro-2 versus GFP-Sec16A vehicle.
Supplementary Dataset 3
Proteomics quantification results GFP-STX5 versus EGFP.
Rights and permissions
About this article
Cite this article
Forrester, A., Rathjen, S.J., Daniela Garcia-Castillo, M. et al. Functional dissection of the retrograde Shiga toxin trafficking inhibitor Retro-2. Nat Chem Biol 16, 327–336 (2020). https://doi.org/10.1038/s41589-020-0474-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0474-4
This article is cited by
-
Kennedy Epitope (KE)-dependent Retrograde Transport of Efficiently Cleaved HIV-1 Envelopes (Envs) and its Effect on Env Cell Surface Expression and Viral Particle Formation
The Protein Journal (2023)
-
Retro-2 alters Golgi structure
Scientific Reports (2022)
-
Halting ErbB-2 isoforms retrograde transport to the nucleus as a new theragnostic approach for triple-negative breast cancer
Cell Death & Disease (2022)
-
Intracellular trafficking of adeno-associated virus (AAV) vectors: challenges and future directions
Gene Therapy (2021)
-
Rendezvous of Retro-2 at the ER
Nature Chemical Biology (2020)