Abstract
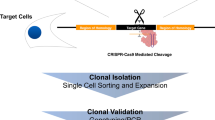
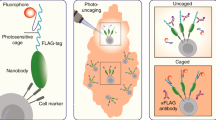
Targeted functional genomics represents a powerful approach for studying gene function in vivo and in vitro. However, its application to gene expression studies in human mast cells has been hampered by low yields of human mast cell cultures and their poor transfection efficiency. We developed an imaging system in which mast cell degranulation can be visualized in single cells subjected to shRNA knockdown or CRISPR–Cas9 gene editing. By using high-resolution confocal microscopy and a fluorochrome-labeled avidin probe, one can directly assess the alteration of functional responses, i.e., degranulation, in single human mast cells (10–12 weeks old). The elimination of a drug or marker selection step avoids the use of potentially toxic treatment procedures, and the brief hands-on time of the functional analysis step enables high-throughput screening of shRNA or CRISPR–Cas9 constructs to identify genes that regulate human mast cell degranulation. The ability to analyze single cells substantially reduces the total number of cells required and enables the parallel visualization of the degranulation profiles of both edited and non-edited mast cells, offering a consistent internal control not found in other protocols. Moreover, our protocol offers a flexible choice between RNA interference (RNAi) and CRISPR–Cas9 genome editing for perturbation of gene expression using our human mast cell single-cell imaging system. Perturbation of gene expression, acquisition of microscopy data and image analysis can be completed within 5 d, requiring only standard laboratory equipment and expertise.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author(s) upon reasonable request; an example dataset is provided as Supplementary Data.
Code availability
The custom Fiji macros described in the main text of this study can be accessed and used by readers without restriction. The code in this manuscript has been peer-reviewed.
References
Galli, S. J. & Tsai, M. IgE and mast cells in allergic disease. Nat. Med 18, 693–704 (2012).
Galli, S. J. et al. Approaches for analyzing the roles of mast cells and their proteases in vivo. Adv. Immunol. 126, 45–127 (2015).
Johnzon, C.-F., Rönnberg, E. & Pejler, G. The role of mast cells in bacterial infection. Am. J. Pathol. 186, 4–14 (2016).
Reber, L. L. et al. Imaging protective mast cells in living mice during severe contact hypersensitivity. JCI Insight 2, 6930 (2017).
Gaudenzio, N. et al. Different activation signals induce distinct mast cell degranulation strategies. J. Clin. Invest 126, 3981–3998 (2016).
Tharp, M. D., Seelig, L. L., Tigelaar, R. E. & Bergstresser, P. R. Conjugated avidin binds to mast cell granules. J. Histochem. Cytochem. 33, 27–32 (1985).
Gaudenzio, N., Laurent, C., Valitutti, S. & Espinosa, E. Human mast cells drive memory CD4+ T cells toward an inflammatory IL-22+ phenotype. J. Allergy Clin. Immunol. 131, 1400–7.e11 (2013).
Joulia, R. et al. Mast cells form antibody-dependent degranulatory synapse for dedicated secretion and defence. Nat. Commun. 6, 6174 (2015).
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J.-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Kleinstiver, B. P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Slaymaker, I. M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Fu, B. X. H., Hansen, L. L., Artiles, K. L., Nonet, M. L. & Fire, A. Z. Landscape of target: guide homology effects on Cas9-mediated cleavage. Nucleic Acids Res. 42, 13778–13787 (2014).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 (2013).
Evers, B. et al. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 34, 631–633 (2016).
Zsebo, K. M. et al. Stem cell factor is encoded at the Sl locus of the mouse and is the ligand for the c-kit tyrosine kinase receptor. Cell 63, 213–224 (1990).
Valent, P. et al. Induction of differentiation of human mast cells from bone marrow and peripheral blood mononuclear cells by recombinant human stem cell factor/kit-ligand in long-term culture. Blood 80, 2237–2245 (1992).
Costa, J. J. et al. Recombinant human stem cell factor (kit ligand) promotes human mast cell and melanocyte hyperplasia and functional activation in vivo. J. Exp. Med 183, 2681–2686 (1996).
Dahlin, J. S. et al. KIT signaling is dispensable for human mast cell progenitor development. Blood 130, 1785–1794 (2017).
Shimizu, Y. et al. Characterization of ‘adult-type’ mast cells derived from human bone marrow CD34+ cells cultured in the presence of stem cell factor and interleukin-6. Interleukin-4 is not required for constitutive expression of CD54, Fc epsilon RI alpha and chymase, and CD13 expression is reduced during differentiation. Clin. Exp. Allergy 32, 872–880 (2002).
Shimizu, Y., Irani, A. M., Brown, E. J., Ashman, L. K. & Schwartz, L. B. Human mast cells derived from fetal liver cells cultured with stem cell factor express a functional CD51/CD61 (alpha v beta 3) integrin. Blood 86, 930–939 (1995).
Andersen, H. B. et al. Comparison of short term in vitro cultured human mast cells from different progenitors—peripheral blood-derived progenitors generate highly mature and functional mast cells. J. Immunol. Methods 336, 166–174 (2008).
Mitsui, H. et al. Development of human mast cells from umbilical cord blood cells by recombinant human and murine c-kit ligand. Proc. Natl Acad. Sci. 90, 735–739 (1993).
Saito, H., Kato, A., Matsumoto, K. & Okayama, Y. Culture of human mast cells from peripheral blood progenitors. Nat. Protoc. 1, 2178–2183 (2006).
Schmetzer, O. et al. A novel method to generate and culture human mast cells: peripheral CD34+ stem cell-derived mast cells (PSCMCs). J. Immunol. Methods 413, 62–68 (2014).
Tam, I. Y. S., Ng, C. W., Tam, S.-Y. & Lau, H. Y. A. Novel six-week protocol for generating functional human connective tissue-type (MCTC) mast cells from buffy coats. Inflamm. Res. 66, 25–37 (2017).
Yin, Y., Bai, Y., Olivera, A., Desai, A. & Metcalfe, D. D. An optimized protocol for the generation and functional analysis of human mast cells from CD34+ enriched cell populations. J. Immunol. Methods 448, 105–111 (2017).
Navinés-Ferrer, A. et al. MRGPRX2-mediated mast cell response to drugs used in perioperative procedures and anaesthesia. Sci. Rep. 8, 11628 (2018).
McNeil, B. D. et al. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature 519, 237–241 (2014).
Hamm, A., Krott, N., Breibach, I., Blindt, R. & Bosserhoff, A. K. Efficient transfection method for primary cells. Tissue Eng. 8, 235–245 (2002).
Joung, J. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Konermann, S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013).
Agrotis, A. & Ketteler, R. A new age in functional genomics using CRISPR/Cas9 in arrayed library screening. Front. Genet. 6, 300 (2015).
de Groot, R., Lüthi, J., Lindsay, H., Holtackers, R. & Pelkmans, L. Large-scale image-based profiling of single-cell phenotypes in arrayed CRISPR-Cas9 gene perturbation screens. Mol. Syst. Biol. 14, e8064 (2018).
Canver, M. C. et al. Integrated design, execution, and analysis of arrayed and pooled CRISPR genome-editing experiments. Nat. Protoc. 13, 946–986 (2018).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015).
Bischoff, S. C. Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nat. Rev. Immunol. 7, 93–104 (2007).
Nilsson, G. et al. Phenotypic characterization of the human mast-cell line HMC-1. Scand. J. Immunol. 39, 489–498 (1994).
Xia, Y. C. et al. Human mast cell line-1 (HMC-1) cells transfected with FcεRIα are sensitive to IgE/antigen-mediated stimulation demonstrating selectivity towards cytokine production. Int. Immunopharmacol. 11, 1002–1011 (2011).
Rådinger, M., Jensen, B. M., Kuehn, H. S., Kirshenbaum, A. & Gilfillan, A. M. Generation, isolation, and maintenance of human mast cells and mast cell lines derived from peripheral blood or cord blood. Curr. Protoc. Immunol. 90, 7.37 (2010).
Guhl, S., Babina, M., Neou, A., Zuberbier, T. & Artuc, M. Mast cell lines HMC-1 and LAD2 in comparison with mature human skin mast cells—drastically reduced levels of tryptase and chymase in mast cell lines. Exp. Dermatol 19, 845–847 (2010).
Laidlaw, T. M. et al. Characterization of a novel human mast cell line that responds to stem cell factor and expresses functional FcεRI. J. Allergy Clin. Immunol. 127, 815–22.e1–5 (2011).
Sheen, C. H., Schleimer, R. P. & Kulka, M. Codeine induces human mast cell chemokine and cytokine production: involvement of G-protein activation. Allergy 62, 532–538 (2007).
Guschin, D. Y. et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 649, 247–256 (2010).
Acknowledgements
We thank C. Liu for technical support and A. van Stigt for helpful discussions regarding MRGPRX2 expression and gene knockout in human mast cell models. We thank P. Dubreuil (Inserm, Marseille, France) for the kind donation of CHO cells. This research was supported by NIH grants U19AI104209, R01 AR067145 and R01 AI32494 (to S.J.G.) and a Marie Skłodowska-Curie Individual Fellowship (H2020-MSCA-IF-2016 no. 749629), the European Research Council (ERC-2018-STG no. 802041), the INSERM ATIP-Avenir program (N.G.) and the Lung Foundation Netherlands (grant 4.1.18.226 to R.W.H). J.F. was supported by a Fulbright Fellowship (financed by the Netherland-America Foundation). R.S. was supported by an NWO Veni Fellowship (grant 91617114) and an Erasmus MC Fellowship.
Author information
Authors and Affiliations
Contributions
J.F. and N.G. designed the experiments and optimized the protocol. J.F. and N.G. performed the experiments and contributed to the data analysis. M.M., R.W.H., R.S., S.-Y.T. and S.J.G. supervised the projects and participated in experimental design and technical discussions. J.F., N.G, M.M., R.W.H., R.S., S.-Y.T. and S.J.G. contributed to the drafting of the manuscript; J.F. wrote the first draft of the manuscript and J.F., S.-Y.T. and S.J.G. completed the finalized version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Hydar Ali and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Gaudenzio, N. et al. J. Clin. Invest. 126, 3981–3998 (2016): https://doi.org/10.1172/JCI85538
Supplementary information
Supplementary Dataset
Supplementary Data and Supplementary Tutorial
Rights and permissions
About this article
Cite this article
Folkerts, J., Gaudenzio, N., Maurer, M. et al. Rapid identification of human mast cell degranulation regulators using functional genomics coupled to high-resolution confocal microscopy. Nat Protoc 15, 1285–1310 (2020). https://doi.org/10.1038/s41596-019-0288-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0288-6
This article is cited by
-
Insights gained from single-cell analysis of chimeric antigen receptor T-cell immunotherapy in cancer
Military Medical Research (2023)
-
Differences and Similarities in the Mechanisms and Clinical Expression of Bradykinin-Mediated vs. Mast Cell–Mediated Angioedema
Clinical Reviews in Allergy & Immunology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.