Abstract
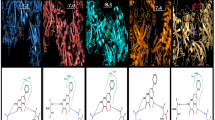
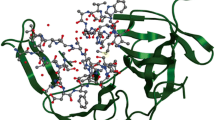
Chikungunya fever has a high morbidity rate in humans and is caused by chikungunya virus (CHIKV). Currently, there is no vaccination or treatment available to show an effective efficacy against this disease. This study targets four non-structural proteins of CHIKV using 650 flavonoids from various medicinal plants, inhabited in Pakistan and India. The compounds are initially screened on the basis of their effective pharmacological properties and are docked against the four proteins. A threshold of − 8.5 kcal/mol is applied to screen and reduce the number of flavonoids for further analysis. The reactivity of screened flavonoids is analyzed using the density functional theory (DFT). Cirsimaritin, apigenin, tamarixetin, and 5,7,3′,4′-tetrahydroxyflavone from Andrographis paniculata have shown a high binding affinity against nsP1. Rhamnetin, tamarixetin and medioresinol have shown a strong binding affinity against nsP2. Four flavonoids, i.e. 5,7,3′,4′-tetrahydroxyflavone, 5,7,4′-trihydroxyflavone, tamarixetin and rhamnetin, showed a high binding affinity for nsP3 while apigenin depicted a strong binding affinity for nsP4. Pharmacological properties of these flavonoids illustrate an effective disposition in humans. The results manifest that the screened eight flavonoids can be analyzed against CHIKV for in vitro and in vivo cell replication, due to their effective pharmacological properties, strong inhibition and high reactivity.







Similar content being viewed by others
References
Mallhi TH, Khan YH, Tanveer N, Bukhsh A, Khan AH, Aftab RA, Khan OH, Khan TM (2018) Awareness and knowledge of Chikungunya infection following its outbreak in Pakistan among health care students and professionals: a nationwide survey. PeerJ 6:e5481
Keramagi AR, Skariyachan S (2018) Prediction of binding potential of natural leads against the prioritized drug targets of chikungunya and dengue viruses by computational screening. 3 Biotech 8(6):274
Oliveira AF, Teixeira RR, Oliveira AS, Souza AP, Silva ML, Paula SO (2017) Potential antivirals: natural products targeting replication enzymes of dengue and chikungunya viruses. Molecules 22(3):505
Rashad AA, Mahalingam S, Keller PA (2013) Chikungunya virus: emerging targets and new opportunities for medicinal chemistry. J Med Chem 57(4):1147–1166
Powers AM (2018) Vaccine and therapeutic options to control chikungunya virus. Clin Microbiol Rev 31(1):e00104–e00116
Al Mahdy A, Jamal M, Kinoshita H, Hossan T (2018) Chikungunya virus outbreak-a threat to global public health including Bangladesh. Bangladesh Journal of Medical Science 17(2):183–184
Lokireddy S, Vemula S, Vadde R (2008) Connective tissue metabolism in chikungunya patients. Virol J 5(1):31
Ji HF, Li XJ, Zhang HY (2009) Natural products and drug discovery. EMBO Rep 10(3):194–200
Mishra K, Sharma N, Diwaker D, Ganju L, Singh S (2013) Plant derived antivirals: a potential source of drug development. J Virol Antivir Res 2:2–9
Delang L, Li C, Tas A, Quérat G, Albulescu I, De Burghgraeve T, Guerrero NS, Gigante A, Piorkowski G, Decroly E (2016) The viral capping enzyme nsP1: a novel target for the inhibition of chikungunya virus infection. Sci Rep 6:31819
Brunetti C, Di Ferdinando M, Fini A, Pollastri S, Tattini M (2013) Flavonoids as antioxidants and developmental regulators: relative significance in plants and humans. Int J Mol Sci 14(2):3540–3555
Hendra R, Ahmad S, Sukari A, Shukor MY, Oskoueian E (2011) Flavonoid analyses and antimicrobial activity of various parts of Phaleria macrocarpa (Scheff.) Boerl fruit. Int J Mol Sci 12(6):3422–3431
Orhan DD, Özçelik B, Özgen S, Ergun F (2010) Antibacterial, antifungal, and antiviral activities of some flavonoids. Microbiol Res 165(6):496–504
Rathee P, Chaudhary H, Rathee S, Rathee D, Kumar V, Kohli K (2009) Mechanism of action of flavonoids as anti-inflammatory agents: a review. Inflamm Allergy Drug Targets 8(3):229–235
Seyedi SS, Shukri M, Hassandarvish P, Oo A, Shankar EM, Abubakar S, Zandi K (2016) Computational approach towards exploring potential anti-chikungunya activity of selected flavonoids. Sci Rep 6:24027
Qaddir I, Rasool N, Hussain W, Mahmood S (2017) Computer-aided analysis of phytochemicals as potential dengue virus inhibitors based on molecular docking, ADMET and DFT studies. J Vector Borne Dis 54(3):255
Rasool N, Iftikhar S, Amir A, Hussain W (2018) Structural and quantum mechanical computations to elucidate the altered binding mechanism of metal and drug with pyrazinamidase from Mycobacterium tuberculosis due to mutagenicity. J Mol Graph Model 80:126–131
Amjad H, Hussain W, Rasool N (2018) Molecular simulation investigation of prolyl oligopeptidase from Pyrobaculum calidifontis and in silico docking with substrates and inhibitors. Open Access J Biomed Eng Biosci 2(4):185–194
Hussain W, Ali M, Sohail Afzal M, Rasool N (2018) Penta-1,4-diene-3-one oxime derivatives strongly inhibit the replicase domain of tobacco mosaic virus: elucidation through molecular docking and density functional theory mechanistic computations. J Antivir Antiretrovir 10(3). https://doi.org/10.4172/1948-5964.1000177
Hussain W, Qaddir I, Mahmood S, Rasool N (2018) In silico targeting of non-structural 4B protein from dengue virus 4 with spiropyrazolopyridone: study of molecular dynamics simulation, ADMET and virtual screening. Virusdisease 29:1–10
Rasool N, Jalal A, Amjad A, Hussain W (2018) Probing the pharmacological parameters, molecular docking and quantum computations of plant derived compounds exhibiting strong inhibitory potential against NS5 from Zika virus. Braz Arch Biol Technol 61. https://doi.org/10.1590/1678-4324-2018180004
Rasool N, Hussain W (2019) Three major phosphoacceptor sites in HIV-1 capsid protein enhances its structural stability and resistance against inhibitor: explication through molecular dynamics simulation, Molecular Docking and DFT Analysis. Comb Chem High Throughput Screen. https://doi.org/10.2174/1386207323666191213142223
Mumtaz A, Ashfaq UA, ul Qamar MT, Anwar F, Gulzar F, Ali MA, Saari N, Pervez MT (2017) MPD3: a useful medicinal plants database for drug designing. Nat Prod Res 31(11):1228–1236
Akhtar A, Amir A, Hussain W, Ghaffar A, Rasool N (2019) In silico computations of selective phytochemicals as potential inhibitors against major biological targets of diabetes mellitus. Curr Comput Aided Drug Des. https://doi.org/10.2174/1573409915666190130164923
Akhtar A, Hussain W, Rasool N (2019) Probing the pharmacological binding properties, and reactivity of selective phytochemicals as potential HIV-1 protease inhibitors. Univ Sci 24(3):441–464
Arif N, Subhani A, Hussain W, Rasool N (2019) In silico inhibition of BACE-1 by selective phytochemicals as novel potential inhibitors: molecular docking and DFT studies. Curr Drug Discov Technol. https://doi.org/10.2174/1570163816666190214161825
Rasool N, Ashraf A, Waseem M, Hussain W, Mahmood S (2019) Computational exploration of antiviral activity of phytochemicals against NS2B/NS3 proteases from dengue virus. Turk J Biochem 44(3): 1–17
Rasool N, Husssain W, Khan YD (2019) Revelation of enzyme activity of mutant pyrazinamidases from Mycobacterium tuberculosis upon binding with various metals using quantum mechanical approach. Comput Biol Chem 83:107108
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9(1):40
Huang B (2009) MetaPocket: a meta approach to improve protein ligand binding site prediction. OMICS 13(4):325–330
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717
Lee SK, Lee IH , Kim HJ, Chang GS, Chung JE, No KT (2003) "The PreADME Approach: Web-based program for rapid prediction of physico-chemical, drug absorption and drug-like properties." EuroQSAR designing drugs and crop protectants: processes, problems and solutions 418–20
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Ai C, Li Y, Wang Y, Li W, Dong P, Ge G, Yang L (2010) Investigation of binding features: effects on the interaction between CYP2A6 and inhibitors. J Comput Chem 31(9):1822–1831
Spessard GO (1998) ACD Labs/LogP dB 3.5 and ChemSketch 3.5. J Chem Inf Comput Sci 38(6):1250–1253
Ghersi D, Sanchez R (2009) Improving accuracy and efficiency of blind protein-ligand docking by focusing on predicted binding sites. Proteins 74(2):417–424
Neese F (2012) The ORCA program system. Wiley Interdiscip Rev Comput Mol Sci 2(1):73–78
Gill PM, Johnson BG, Pople JA, Frisch MJ (1992) The performance of the Becke—Lee—Yang—Parr (B—LYP) density functional theory with various basis sets. Chem Phys Lett 197(4–5):499–505
Lovell SC, Davis IW, Arendall III WB, De Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins: Structure, Function, and Bioinformatics 50(3):437–450
Nguyen PT, Yu H, Keller PA (2015) Identification of chikungunya virus nsP2 protease inhibitors using structure-base approaches. J Mol Graph Model 57:1–8
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23(1–3):3–25
Tran N (2011) Blood-brain barrier. Encycl Clin Neuropsychol:426–426
Ritter J (2008) Wiley handbook of current and emerging drug therapies. Br J Clin Pharmacol 65(3):449
Szymański P, Markowicz M, Mikiciuk-Olasik E (2012) Adaptation of high-throughput screening in drug discovery—toxicological screening tests. Int J Mol Sci 13(1):427–452
Parashar D, Cherian S (2014) Antiviral perspectives for chikungunya virus. Biomed Res Int 2014:631642
Oo A, Hassandarvish P, Chin SP, Lee VS, Bakar SA, Zandi K (2016) In silico study on anti-Chikungunya virus activity of hesperetin. PeerJ 4:e2602
Ahmadi A, Hassandarvish P, Lani R, Yadollahi P, Jokar A, Bakar SA, Zandi K (2016) Inhibition of chikungunya virus replication by hesperetin and naringenin. RSC Adv 6(73):69421–69430
Mishra P, Kumar A, Mamidi P, Kumar S, Basantray I, Saswat T, Das I, Nayak TK, Chattopadhyay S, Subudhi BB (2016) Inhibition of chikungunya virus replication by 1-[(2-methylbenzimidazol-1-yl) methyl]-2-oxo-indolin-3-ylidene] amino] thiourea (MBZM-N-IBT). Sci Rep 6:20122
Eroglu E, Türkmen H (2007) A DFT-based quantum theoretic QSAR study of aromatic and heterocyclic sulfonamides as carbonic anhydrase inhibitors against isozyme, CA-II. J Mol Graph Model 26(4):701–708
Gogoi D, Baruah VJ, Chaliha AK, Kakoti BB, Sarma D, Buragohain AK (2017) Identification of novel human renin inhibitors through a combined approach of pharmacophore modelling, molecular DFT analysis and in silico screening. Comput Biol Chem 69:28–40
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
The authors declare that no human participants or animals were involved in this study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 20 kb)
Rights and permissions
About this article
Cite this article
Hussain, W., Amir, A. & Rasool, N. Computer-aided study of selective flavonoids against chikungunya virus replication using molecular docking and DFT-based approach. Struct Chem 31, 1363–1374 (2020). https://doi.org/10.1007/s11224-020-01507-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-020-01507-x