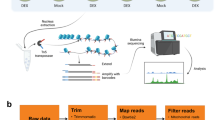
Abstract
Eukaryotic gene transcription is associated with the eviction of nucleosomes and the formation of open chromatin, which enables the recruitment of transcriptional coactivators and other regulatory factors. Open chromatin is thus a hallmark of functional regulatory DNA elements in genomes. In recent years, formaldehyde-assisted isolation of regulatory elements (FAIRE) has proven powerful in identifying open chromatin in the genome of various eukaryotes, particularly yeast, human, and mouse. However, it has proven challenging to adapt the FAIRE protocol for use on plant material, and the few available protocols all have their drawbacks (e.g., applicability only to specific developmental stages). In this Protocol Extension, we describe a reliable FAIRE protocol for mature Arabidopsis (Arabidopsis thaliana) leaves that adapts the original protocol for use on plants. The main differences between this protocol extension and the earlier FAIRE protocol are an increased formaldehyde concentration in the chromatin crosslinking buffer, application of a repeated vacuum to increase crosslinking efficiency, and altered composition of the DNA extraction buffer. The protocol is applicable to leaf chromatin of unstressed and stressed plants and can be completed within 1 week. Here, we also describe downstream analysis using qPCR and next-generation sequencing. However, this Protocol Extension should also be compatible with downstream hybridization to a DNA microarray. In addition, it is likely that only minor adaptations will be necessary to apply this protocol to other Arabidopsis organs or plant species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The unprocessed data of this study are available from the corresponding author upon reasonable request.
References
Wallrath, L. L., Lu, Q., Granok, H. & Elgin, S. C. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays 16, 165–170 (1994).
Boyle, A. P. et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 21, 456–464 (2011).
Boeger, H., Griesenbeck, J., Strattan, J. S. & Kornberg, R. D. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11, 1587–1598 (2003).
Tsompana, M. & Buck, M. J. Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7, 33 (2014).
Nagy, P. L., Cleary, M. L., Brown, P. O. & Lieb, J. D. Genome-wide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc. Natl. Acad. Sci. USA 100, 6364–6369 (2003).
Lee, K. et al. Genetic landscape of open chromatin in yeast. PLoS Genet. 9, e1003229 (2013).
Ponts, N. et al. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 20, 228–238 (2010).
Strenkert, D., Schmollinger, S., Sommer, F., Schulz-Raffelt, M. & Schroda, M. Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell 23, 2285–2301 (2011).
Giresi, P. G., Kim, J., McDaniell, R. M., Iyer, V. R. & Lieb, J. D. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 17, 877–885 (2007).
Giresi, P. G. & Lieb, J. D. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements). Methods 48, 233–239 (2009).
Gaulton, K. J. et al. A map of open chromatin in human pancreatic islets. Nat. Genet. 42, 255–259 (2010).
Simon, J. M., Giresi, P. G., Davis, I. J. & Lieb, J. D. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 7, 256–267 (2012).
Louwers, M. et al. Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell 21, 832–842 (2009).
Omidbakhshfard, M. A., Winck, F. V., Arvidsson, S., Riaño-Pachón, D. M. & Mueller-Roeber, B. A step-by-step protocol for formaldehyde-assisted isolation of regulatory elements from Arabidopsis thaliana. J. Integr. Plant Biol. 56, 527–538 (2014).
Baum, S. et al. Isolation of open chromatin identifies regulators of systemic acquired resistance in Arabidopsis. Plant Physiol. 181, 817–833 (2019).
Schillheim, B. et al. Sulforaphane modifies histone H3, unpacks chromatin, and primes defense in Arabidopsis. Plant Physiol. 176, 2395–2405 (2018).
Song, L. et al. Open chromatin defined by DNase I and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 21, 1757–1767 (2011).
Blankenship, R. E. & Hartman, H. The origin and evolution of oxygenic photosynthesis. Trends Biochem. Sci. 23, 94–97 (1998).
Perduns, R., Horst-Niessen, I. & Peterhaensel, C. Photosynthetic genes and genes associated with the C4 trait in maize are characterized by a unique class of highly regulated histone acetylation peaks on upstream promoters. Plant Physiol. 168, 1378–1388 (2015).
Zhang, W. & Jiang, J. Genome-wide mapping of DNase I hypersensitive sites in plants. Methods Mol. Biol. 1284, 71–89 (2015).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Buenrosto, J., Wu, B., Chang, H. & Greenleaf, W. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2016).
Bajic, M., Maher, K. A. & Deal, R. B. Identification of open chromatin regions in plant genomes using ATAC-seq. Methods Mol. Biol. 1675, 183–201 (2018).
Maher, K. A. et al. Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell 30, 15–36 (2018).
ENCODE Project Consortium. A user’s guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol. 9, e1001046 (2011).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Berardini, T. Z. et al. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Fu, Z. Q. & Dong, X. Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863 (2013).
Van Loon, L. C., Rep, M. & Pieterse, C. M. J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162 (2006).
Takahashi, T. & Komeda, Y. Characterization of two genes encoding small heat-shock proteins in Arabidopsis thaliana. Mol. Gen. Genet. 219, 365–372 (1989).
Kodama, Y., Nagaya, S., Shinmyo, A. & Kato, K. Mapping and characterization of DNase I hypersensitive sites in Arabidopsis chromatin. Plant Cell Physiol. 48, 459–470 (2007).
Acknowledgements
We thank S.F. Beyer for help with the figures. U.C. was supported by a grant from the German Research Foundation (CO 186/13-1) and by the Excellence Initiative of the German federal and state governments.
Author information
Authors and Affiliations
Contributions
U.C. initiated and supervised the project. M.R.J. introduced S.B. to chromatin techniques. S.B. and E.-M.R.-M. performed the experiments. U.C. wrote the article with contributions from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Jeremy M. Simon and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference(s) using this protocol
Baum, S. et al. Plant Physiol. 181, 817–833 (2019): https://doi.org/10.1104/pp.19.00673
Schillheim, B. et al. Plant Physiol. 176, 2395–2405 (2018): https://doi.org/10.1104/pp.17.00124
This protocol is an extension to: Nat. Protoc. 7, 256–267 (2012): https://doi.org/10.1038/nprot.2011.444
Integrated supplementary information
Supplementary Figure 1 Open chromatin in the 5′ regulatory region and around the TSS of the PR1 gene in systemic leaves relative to the UBIQUITIN5 promoter.
The experiment was done three times (n = 3). ***, P = 0.001; **, P = 0.01. Measurements were taken from distinct samples. Normal distribution was assumed for all statistical analyses. Unpaired Student´s t tests (two-sided) was applied using SigmaStat 4.0 (Systat Software, Inc.) to determine whether the observed differences were statistically significant. Changes were considered statistically significant when P ≤ 0.01. Psm, Pseudomonas syringae pv. maculicola; TSS, transcription start site; bp, base pairs; R1 - R3, biological replicates 1–3.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Baum, S., Reimer-Michalski, EM., Jaskiewicz, M.R. et al. Formaldehyde-assisted isolation of regulatory DNA elements from Arabidopsis leaves. Nat Protoc 15, 713–733 (2020). https://doi.org/10.1038/s41596-019-0277-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0277-9
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.