Abstract
Hornworts, liverworts and mosses are three early diverging clades of land plants, and together comprise the bryophytes. Here, we report the draft genome sequence of the hornwort Anthoceros angustus. Phylogenomic inferences confirm the monophyly of bryophytes, with hornworts sister to liverworts and mosses. The simple morphology of hornworts correlates with low genetic redundancy in plant body plan, while the basic transcriptional regulation toolkit for plant development has already been established in this early land plant lineage. Although the Anthoceros genome is small and characterized by minimal redundancy, expansions are observed in gene families related to RNA editing, UV protection and desiccation tolerance. The genome of A. angustus bears the signatures of horizontally transferred genes from bacteria and fungi, in particular of genes operating in stress-response and metabolic pathways. Our study provides insight into the unique features of hornworts and their molecular adaptations to live on land.
Similar content being viewed by others
Main
Land plants (Embryophyta) probably originated in the early Palaeozoic1, initiating the colonization of the terrestrial habitat. Because bryophytes (hornworts, liverworts and mosses) emerged from the early split in the diversification of land plants, they are key to the study of early land plant evolution (Supplementary Note 1.1). Unlike other extant land plants, the vegetative body of bryophytes is the haploid gametophyte, the sporophyte is always unbranched and permanently attached to the maternal plant, and both generations lack lignified vascular tissue2. Bryophytes occur in nearly all terrestrial habitats on all continents but are absent from marine environments3.
With only 200–250 species worldwide, the diversity of hornworts is much lower than that of the other six extant lineages of embryophytes (angiosperms, gymnosperms, ferns, lycophytes, mosses and liverworts)4. Long considered sister to all other land plants, or sister to all extant vascular plants, hornworts have recently been resolved as sister to the setaphytes (that is, the mosses and liverworts) within monophyletic bryophytes1,5,6,7,8. Still, hornworts possess a series of distinct features9. For instance, most hornworts have chloroplasts with CO2-concentrating pyrenoids, which have not been found in any other land plants but are widespread among green algae10. Other unusual features of hornworts include the persistent basal meristem in the sporophyte and mucilage-filled cavities for colonial symbionts on the gametophyte11. Most hornworts form tight symbiotic relationships with cyanobacteria12 and fungal endophytes (especially Glomeromycota and Mucoromycotina)13.
Here, we present the draft genome of A. angustus Steph. (Anthocerotaceae) (see Methods, Supplementary Figs. 1 and 2, and Supplementary Note 1.2). Completion of this high-quality hornwort genome complements previously sequenced representatives of the mosses (Physcomitrella patens14) and liverworts (Marchantia polymorpha15) and provides a unique opportunity to revisit bryophyte phylogeny, early land plant evolution and the adaptation of plants to live on land.
Genome assembly and annotation
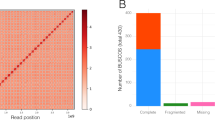
We sequenced the genome of A. angustus (a single individual of unknown sex from the dioecious species) using a combination of Illumina and Oxford Nanopore high-throughput sequencing systems (see Methods). We generated 126.53 Gb raw reads from Illumina and 63.61 Gb raw reads from Nanopore sequencing platforms, and retained 17.10 Gb and 3.78 Gb, respectively, after filtering, error-correction and decontamination (see Methods, Supplementary Figs. 2–4 and Supplementary Tables 1–3). Finally, we obtained an optimized assembly of 119 Mb with a contig N50 length of 796.64 kb and a scaffold N50 length of 1.09 Mb (Table 1 and Supplementary Table 4). Approximately 97.66% of the vegetative gametophyte transcriptome data for A. angustus genome annotation can be mapped to the assembled genome (Supplementary Table 5). Repeat sequences comprise 64.21% of the assembled genome, with transposable elements (TEs) being the major component (Table 1 and Supplementary Tables 6 and 7). Among the TEs, long terminal repeats (LTRs) are the most abundant (Supplementary Table 7). We used a combination of de novo, homology-based and RNA sequence-based predictions to obtain gene models for the A. angustus genome (Supplementary Table 8). In total, we predicted 14,629 protein-coding genes with an average coding-sequence length of 1.31 kb and an average of 4.81 exons per gene (Table 1, Supplementary Fig. 5 and Supplementary Table 8). About 85% of these predicted genes have their best hits on plant sequences from the National Center for Biotechnology Information (NCBI) non-redundant database (Supplementary Fig. 6), and 78.39% were functionally annotated through Swissprot, TrEMBL, Pfam, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Supplementary Tables 9 and 32). Our annotation captured 89.64% of the 956 genes in the BUSCO plantae dataset16 (85.04% complete gene models plus 4.60% fragmented gene models), compared with 93.51% and 92.15% captured in P. patens14 and M. polymorpha15, respectively (Supplementary Table 10). In addition to protein-coding genes, we also identified 30 known mature micro RNAs (miRNAs), 180 novel mature miRNAs, 347 transfer RNAs, 94 ribosomal RNAs and 83 small nuclear RNAs (snRNAs) in the A. angustus genome (Supplementary Table 11). Nine mature miRNA sequences that appear conserved among land plants (miR156/157, miR159/319, miR160, miR165/166, miR170/171, miR408, miR477, miR535 and miR536)17 were also found in A. angustus (Supplementary Table 12).
Comparative genomic analysis
For sequence similarity-based clustering of homologues, we used the predicted proteomes of A. angustus and 18 other green plants with fully-sequenced genomes (that is, 11 other land plants, two charophyte green algae and five chlorophyte green algae; Supplementary Table 13). Genes of A. angustus are distributed among 7,644 gene families that are shared with other plants, and 497 gene families that appear to be unique to A. angustus (Fig. 1a and Supplementary Table 14). In the shared gene families, most A. angustus genes (that is, 9,680) cluster with land plant genes, and only a very small number (that is, 107) specifically cluster with green algae genes (Supplementary Fig. 7). The gene families unique to A. angustus are enriched in various biosynthetic categories (for example, terpenoid and zeatin) and various activity categories (for example, nutrient reservoir activity and catechol oxidase activity) (Supplementary Tables 15 and 16).
a, Comparison of the number of gene families identified by OrthoMCL. The Venn diagram shows the shared and unique gene families in A. angustus, Setaphyta, Tracheophyta, Charophyta and Chlorophyta. The gene-family number is listed in each of the components. b, Gene-family gain (+)/loss (−) among 19 green plants. The numbers of gained (blue) and lost (red) gene families are shown above the branches. The boxed number indicates the gene-family size at each node. The number of gene families, orphans (single-copy gene families) and number of predicted genes is indicated next to each species. c, Comparison of whole paranome, anchor pair and one-to-one orthologue distribution of the number of synonymous substitutions per synonymous site (KS) across the three bryophyte species (P. patens, M. polymorpha and A. angustus).
Phylogenetic inferences from 85 single-copy nuclear genes sampled for A. angustus and 18 other green plants resolve hornworts (A. angustus), mosses (P. patens) and liverworts (M. polymorpha) as a monophyletic group, with hornworts sister to mosses and liverworts, which agrees with inferences from 852 nuclear genes sampled from 103 plant species1 (Fig. 1b, Supplementary Figs. 8 and 9, Supplementary Table 17 and Supplementary Notes 2.1 and 2.2). The divergence (Supplementary Figs. 10 and 11, Supplementary Tables 18–20 and Supplementary Note S2.3) of the extant crown group of hornworts is estimated at 275.62 million years ago (Ma) (95% highest posterior density, 179.3–384.6 Ma) (middle Carboniferous–early Jurassic) (Supplementary Fig. 11 and Supplementary Table 20), which is comparable to the crown age of hornworts estimated based on two organellar sequences from 77 hornworts and 11 other land plants10. These estimates are thus older than those inferred from the fossil record, considering that the oldest putative hornwort fossil is a spore from the Lower Cretaceous Baqueró Formation, Argentina (from 145 to 100 Ma) that resembles the spores of extant Anthoceros18.
Comparative genomics shows that the genome of A. angustus has lost many gene families (that is, 2,145) and comparatively only modest gains (that is, 497) (Fig. 1b). A similar trend characterizes the genome of Marchantia and of the ancestor common to all bryophytes, whereas P. patens has gained more families (that is, 1,334) than it has lost (that is, 1,248; Fig. 1b). Thus, bryophyte genomes may not only harbour a number of genes and gene families comparable to those of vascular plants and in particular seed plants (Fig. 1b) but may also be highly dynamic through evolutionary time.
Many, if not most, land plants harbour genomic signatures of ancient whole-genome duplication (WGD)19. However, like that of Marchantia15, the genome of Anthoceros lacks evidence of having undergone a WGD (Fig. 1c, Supplementary Fig. 13 and Supplementary Note 3.1), which confirms the hypothesis drawn previously from the analysis of transcriptomic data20. The chromosomal arrangement of genes is not much conserved among the three bryophyte lineages (Supplementary Fig. 14a,b and Supplementary Note 3.2), which likely reflects the ancient divergence of these different lineages of bryophytes. For example, the longest co-linear block corresponds to a mere five anchor pairs for both A. angustus versus P. patens and A. angustus versus M. polymorpha, whereas within the A. angustus genome, the largest co-linear segment consists of six anchor pairs (Supplementary Fig. 14).
The A. angustus genome contains a much lower percentage of multi-copy gene families than that of single-copy gene families, implying low genetic redundancy (Supplementary Table 17), which is similar to what has been observed for the liverwort Marchantia15.
Transcription factors
The A. angustus genome comprises 333 putative transcription factor (TF) genes covering 61 families, a number that is highly similar to that of the other two bryophyte genomes (Supplementary Fig. 15, Supplementary Table 21 and Supplementary Note 4.1). The diversity of TF genes in extant plants is rather stable (Supplementary Fig. 15) and resulted from two ancient bursts of TF families during the diversification of green plants: one concomitant with the origin of streptophytes and the other with the transition to land15,21. In plants, genes encoding TFs are among the most highly retained following polyploidy22, a pattern reflected in the comparison of the three bryophyte genomes14,15. A. angustus and M. polymorpha, whose genome did not undergo WGDs hold a small number of TF compared to P. patens, which experienced at least one WGD in its ancestry, resulting in a substantially larger number of TF genes (Supplementary Fig. 15). It supports the hypothesis that the WGD is an important mechanism for expansion of TF families23.
Phylogenetic analyses of 24 gene families contributing to the development of plant body plans or adaptation to the terrestrial environment, including 16 TF gene families24,25 (Fig. 2a, Supplementary Figs. 16–54, Supplementary Table 22 and Supplementary Note 4.2), confirm that a considerable number of genes, such as genes involved in gametophyte or sporophyte development, haploid–diploid transition, meristem development, filamentous growth, photomorphogenesis and auxin signalling (Fig. 2), composed the genetic toolkit of plants before the conquest of land26. In particular, the TF genes for filamentous growth and auxin signalling arose in charophyte green algae27,28 (Fig. 2b), which are thought to be the closest living relatives to extant land plants, implying the preliminary establishment of relatively more complex body plan in these basal streptophytes for plant terrestrial adaptation29. Furthermore, a set of genes underlying key morphological innovations for terrestrial adaptation probably evolved along with the colonization of land30,31 (Fig. 2b), such as SMF and ICE for stomatal development (Supplementary Figs. 29 and 30), APB, CLE and CLV1 for 3D growth (Supplementary Figs. 36 and 50–52), and VNS for water-conducting-cell development (Supplementary Fig. 38). The sporophyte morphology of bryophytes is relatively simple, and many of the genes involved in the elaborate regulation of embryogenesis32, such as FUS3, LEC1, LEC2, NF-YA1/9 and NF-YA3/5/6/8 are absent in A. angustus, Marchantia and Physcomitrella (Fig. 2a and Supplementary Figs. 39–41). The ABI3 genes that mainly function in embryo maturation and seed desiccation tolerance in flowering plants are present in bryophytes, and have roles in desiccation tolerance in their vegetative tissues33.
In A. angustus, most genes involved in the development of plant body plans have a single copy, and a few A. angustus TF gene families even lost a subset of duplicates (Fig. 2a and Supplementary Figs. 16–52). For example, in the bHLH family, the class I RSL gene that controls the development of rhizoids and root hairs, thought to have been important for the colonization of land34, is present in the A. angustus genome, whereas the class II RSL genes responsible for regulating protonema differentiation in P. patens or root hair elongation in A. thaliana by auxin35 are absent (Supplementary Fig. 27 and Supplementary Note 4.2). The lack of class II RSL genes in A. angustus might be related to the morphological simplification of this species with respect to tip-growing filamentous structures2. For the KNOX genes from the homeobox gene family, the A. angustus genome retains one class II KNOX gene for haploid-to-diploid morphological transition36, but lacks class I KNOX genes (Supplementary Fig. 23), whose activity is necessary for seta extension in the sporophytes in P. patens37. The absence of this gene might be linked to the absence of setae in hornworts2. The genome of A. angustus also holds few type II MIKCC MADS-box, class B ARF, NCARF and short PIN genes, as a result of gene losses suggested by our phylogenetic analysis (Supplementary Figs. 17, 42, 45 and Supplementary Note 4.2). The class II RSL, class B ARF, NCARF and short PIN genes all have auxin-related functions (Supplementary Note 4.2). Since these auxin-related genes were consistently lost in A. angustus, this hornwort species possesses the simplest auxin molecular toolkit among all investigated land plants so far38. Thus, like the liverwort M. polymorpha15, A. angustus exhibits low redundancy for genes shaping the plant body plan (Fig. 2b). Such a limited toolkit may be characteristic of the ancestor to bryophytes and hence, perhaps, of the earliest land plants with a dominant thalloid gametophyte, and provide the foundation to explaining the architectural simplicity of these plants. By contrast, the genome of P. patens, which develops a leafy stem, has the most TF genes involved in the development of plant body plans among the compared bryophytes (Fig. 2b). Although the genome of A. angustus seems poor in genes composing the network underlying the development of its body plan, the TF gene families linked to responses to terrestrial environmental stimuli exhibit lineage-specific gene expansions in A. angustus, namely, the LISCL genes for mycorrhizal signalling in the GRAS gene family39 (Supplementary Fig. 53) and the clade SIP1 for ABA signalling under water stress in the Trihelix gene family40 (Supplementary Fig. 54).
Gene-family expansion
Besides two TF gene families, the A. angustus genome harbours a variety of other uniquely expanded gene families (Supplementary Fig. 55). The genome comprises an very large number of pentatricopeptide repeat (PPR) genes for plant organellar RNA processing41, accounting for approximately 7.90% of the predicted protein-coding genes. The expanded PPR genes are PLS-class PPR genes (Supplementary Fig. 55, Supplementary Tables 23 and 24 and Supplementary Note 5.1). Most of the PLS-class PPR proteins in A. angustus were predicted to be localized in the mitochondrion or chloroplast (Supplementary Table 24). The expansion of the PLS-class PPR genes correlates with the large number of RNA editing sites estimated in the organellar genomes of A. angustus (Supplementary Table 23). Our findings add further support to the hypothesis that an increase in the number of both RNA editing sites and PPR genes (especially the PLS-class PPR) occurred after the separation of land plants from green algae41,42 (Supplementary Table 23). The reduced number of PPR genes and absence of RNA editing in marchantiid liverworts are most probably secondary losses (Supplementary Table 23), as the organellar RNA editing and plant-specific extensions of PPR genes were also found in jungermanniid liverworts43. Through RNA editing, the PPR proteins could act as ‘repair’ factors that alleviate DNA damage caused by increased UV exposure in terrestrial environments41. Other stress-response gene families have also expanded in A. angustus, such as cupin and cytochrome P450 (CYP) (Supplementary Fig. 55). Two groups of cupin (PF00190) proteins—that is, monocupins and bicupins—can be recognized on the basis of the number of cupin domains44. In A. angustus, the cupin gene family has undergone a significant expansion (Supplementary Table 25) such that it comprises more bicupin genes than any other plant (Fig. 3a, Supplementary Figs. 56 and 57, Supplementary Table 25 and Supplementary Note 5.2). Expansion of the cupin gene family in A. angustus resulted mainly from tandem gene duplications (Fig. 3b,c and Supplementary Note 5.2). Since bicupins (that is, 11S and 7S seed storage proteins) are desiccation-tolerant proteins in higher land plants44, the large number of bicupin genes in A. angustus could indicate adaptation for coping with drought stress in the terrestrial environment. The large number of A. angustus-specific monocupin genes are homologous to the P. patens PpGLP6 gene (XP_001782709.1) (Supplementary Fig. 57 and Supplementary Note 5.2), which encodes a protein with manganese-containing extracellular superoxide dismutase (SOD) activity to respond to oxidative stress in terrestrial environments45. The CYP genes for primary and secondary metabolism have also expanded in A. angustus (Supplementary Fig. 55 and Supplementary Note 5.3). For instance, genes belonging to the subfamilies CYP71 and CYP85 contain 56 and 46 genes, respectively (Supplementary Figs. 58–61 and Supplementary Tables 26 and 27). The A. angustus CYP genes were assigned to 28 KEGG pathways, of which ‘flavonoid 3′-monooxygenase/flavonoid 3′,5′-hydroxylase’ and ‘abscisic acid 8′-hydroxylase’ were the most representative (Supplementary Table 28). Within the CYP71 gene subfamily, genes homologous to flavonoid 3′-hydroxylase (monooxygenase) (F3'H) or flavonoid 3′,5′-hydroxylase (F3′5′H) genes that are involved in flavonoid biosynthesis46 are highly expanded in A. angustus (Supplementary Fig. 59 and Supplementary Note 5.3). Because flavonoids have an important role in UV-B protection46, the expansion of flavonoid biosynthesis related genes in A. angustus might again represent a molecular adaptation to life in the terrestrial environment. Among the CYP85 genes, the genes homologous to abscisic acid 8′-hydroxylase genes involved in abscisic acid catabolism during drought stress response47 are also uniquely abundant in A. angustus (Supplementary Fig. 60 and Supplementary Note 5.3), and may account for the high desiccation tolerance of A. angustus. Like the cupin gene family, many of the above expanded gene families occur in tandem arrays (Supplementary Table 29). At least 9.82% of protein-coding genes in A. angustus form ‘tandem’ clusters in the genome (Supplementary Table 30 and Supplementary Note 5.4), compared with only 1% in P. patens14 and 5.9% in M. polymorpha15.
a, A summary of the number of cupin genes from nine species based on a Pfam search of cupin_1 domain (PF00190). b,c, Phylogenetic trees show cupin genes in nine plant genomes: bicupins (b) and monocupins (c). The colour of each branch corresponds to the background colour for each species in a. The tandem duplicated gene clusters are ordered and shown on scaffolds of the A. angustus genome. The scale bars in the trees show the number of amino acid substitutions per site.
CO2-concentrating mechanism
Hornworts are the only extant land plant lineage harbouring a pyrenoid-based CO2-concentrating mechanism (CCM) similar to that of green algae9,48 (Supplementary Note 6.1), for which the key components have been identified49. To clarify whether the CCM components of green algae have orthologues in hornworts and other land plants, we searched the A. angustus genome and other plant genomes or transcriptomes with reference to the CCM genes from chlorophyte green algae Chlamydomonas reinhardtii49,50 (Supplementary Figs. 62–71 and Supplementary Note 6.2). A. angustus and all other green plants harbour orthologues of CAH1/2 whose expression is modulated by external inorganic carbon concentration; of CemA, which maintains stromal pH balance; of LCI11, which mediates the entry of HCO3− in the thylakoid lumen; and of RCA1 and RBCS1/2, which regulate CO2 fixation by Rubisco (Supplementary Figs. 62, 65 and 69–72). By contrast, orthologues of CCP1/2, which mediate the entry of HCO3- into the chloroplast stroma and of EPYC1, which regulate CO2 fixation by Rubisco were only present in chlorophyte green algae (Supplementary Figs. 67 and 72 and Supplementary Note 6.2). The three inorganic carbon transporters (HLA3, LCI1 and LCIA-like genes) only occur in bryophytes and green algae, whereas the A. angustus genome lacks the related orthologues (Supplementary Figs. 63, 66 and 72 and Supplementary Note 6.2). Unexpectedly, the three kinds of carbonic anhydrases (CAH3, CAH9 and LCIB/C), which are essential components of CCM, are conserved in non-angiosperm land plants and green algae (Supplementary Figs. 62, 64, 68 and 72). The A. angustus genome retains the orthologues of both LCIB/C and CAH3 genes, but has no copy of CAH9 (Supplementary Fig. 72). Besides green algae, the essential CCM components occur in both hornworts and other non-angiosperm land plants that lack pyrenoids (Supplementary Fig. 72). It implies that the CCM could be an ancestral mechanism of CO2 fixation by plants, and pyrenoids for CCM are homologous between hornworts and green algae, whereas both CCM components and pyrenoids have undergone multiple losses in land plants in response to atmospheric changes in terrestrial environments10,48.
Horizontal gene transfer
Horizontal gene transfer (HGT) from bacteria or fungi has been reported for both the moss P. patens51 and the liverwort M. polymorpha15. Consistent with those observations, the taxonomic distribution of BLASTP hits following careful phylogenetic analysis and manual inspection suggested that 19 genes from 14 families originated from HGTs from either bacteria or fungi (Supplementary Fig. 6 and Supplementary Note 7.1). Bacterial donors are distributed among nine families: Actinobacteria (three gene families), Alphaproteobacteria (two gene families), Bacteroidetes (two gene families), Firmicutes (one gene family) and Verrucomicrobia (one gene family). Five families were acquired from fungi, belonging to Ascomycota, Basidiomycota, hornwort-symbiotic Chytridiomycota or Mucoromycota13 (Fig. 4a,b, Supplementary Figs. 73–84 and 86 and Supplementary Table 31). The detection of specific HGT in all three fully sequenced bryophytes is remarkable, and is probably related to the fact that these organisms form symbioses with diverse bacteria and fungi, which, together with the weakly protected tissues in the early developmental stages in the life cycle of these plants, provide the possibility for HGT51. In addition, we found that two families originating from HGT from bacteria are shared by the three bryophyte lineages, and one originating from a HGT from fungi is shared between hornworts and liverworts only (Fig. 4c,d, Supplementary Figs. 85 and 86, Supplementary Table 31 and Supplementary Note 7.2). The HGT genes mentioned above (SCUO value 0.2127) exhibit a significantly more biased codon-usage pattern than non-HGT genes (SCUO value 0.1595) (Supplementary Fig. 87a), which may be linked to their higher GC content (57.58%) than non-HGT genes (53.26%) (Supplementary Fig. 87b).
a, Phylogenetic tree of glyoxalase (PF13468). b, Phylogenetic tree of NAD-binding dehydrogenase (PF08635). c, Phylogenetic tree of glucuronyl hydrolase (PF07470). d, Phylogenetic tree of DNA methyltransferase (PF02870 and PF01035). The stars indicate that the Anthoceros sequence or bryophyte sequences formed a monophyletic clade with homologues of putative HGT donor, reflecting Anthoceros-specific or bryophyte-specific HGT events. Maximum-likelihood bootstrap support values ≥50% are shown above the branches. Red, hornworts and other bryophytes; cyan, green algae; grey, metazoan; orange, stramenopiles; blue, bacteria; yellow, fungi; purple, archaea. The homologues from the kingdom other than the one that HGT donors are involved in are used as the outgroup. The scale bars in the trees show the number of amino acid substitutions per site.
The HGT-derived genes in A. angustus mainly contribute to metabolic processes, oxidation–reduction and stress response (Supplementary Table 31). Some transferred genes related to carbohydrate metabolism are predicted to encode glucuronyl (AANG011893) and glycosyl hydrolases (AANG004297) (Fig. 4c, Supplementary Fig. 79 and Supplementary Table 31), which function in cell wall synthesis and modification and might extend the metabolic flexibility of A. angustus in changing environments52. The Alphaproteobacteria-derived gene AANG004679 encodes glyoxalase, which is related to drought stress tolerance53 (Fig. 4a). The Actinobacteria-derived DNA methyltransferase genes that are present only in the three groups of bryophytes are related to DNA repair54 (Fig. 4d). The hornworts and liverworts share the fungi-derived terpene synthase-like (MTPSL) genes (Supplementary Fig. 85). Terpene synthases are pivotal enzymes for the biosynthesis of terpenoids, which serve as chemical defences against herbivores and pathogens55. Some horizontally transferred genes in A. angustus, such as NAD-binding dehydrogenase (Fig. 4b) and MTPSL genes (Supplementary Fig. 85), underwent subsequent gene duplications. The results suggest that the acquisition of foreign genes might have provided additional means for environmental adaptation during evolution of the hornwort lineage.
Conclusions
As land pioneers, the three bryophyte groups form a well-supported monophyletic lineage, with hornworts sister to liverworts and mosses. The genome of hornwort A. angustus shows no evidence of WGDs and low genetic redundancy for networks underlying plant body plan, which may be congruent with an overall simple body plan. Hornworts have retained the essential components of CCM found in green algae in response to the atmospheric changes in terrestrial environments. Meanwhile, the gene inventory in A. angustus expanded mainly through tandem duplication and HGT. In particular, the expansion of specific gene families and the acquisition of foreign genes have provided additional metabolic abilities in hornworts that probably facilitated their survival in a terrestrial environment. Together, our results indicate how the draft genome of A. angustus provides a useful model for studying early land plant evolution and the mechanism of plant terrestrial adaptation.
Methods
Sample preparation and sequencing
The natural populations of A. angustus Steph. were collected from Jinping County, Yunnan Province, China. The voucher specimen has been deposited at the herbarium, Institute of Botany, Chinese Academy of Sciences, Beijing, China with collection number W1879-2010-01-18. The sporophytes of A. angustus were detached from the gametophytes, sterilized in 10% sodium hypochlorite and subsequently rinsed with distilled water56. The sporangium was opened and the spores were homogenized and spread onto the 1/2 KnopII agar medium57 in Petri dishes (Supplementary Fig. 1b). The culture temperature was between 21 °C and 25 °C. Spores germinated within a couple of days, and then the sporelings started to grow. After approximately three to four weeks, the gametophyte started to grow (Supplementary Fig. 1c,d). Since spores are aposymbiotic, we did not find the phenomenon of mucilage-filled cavities colonization by cyanobacteria on the A. angustus gametophyte during the sterile culture. A gametophyte from a single spore was selected and cultured by asexual propagation. The tissue yielded from subculture was used for genome and RNA sequencing. We tried to induce sexual reproduction by dropping the growth temperature of gametophyte cultures to 10 °C and 16 °C, respectively; however until now they have not yet produced reproductive organs. Therefore, the sequenced A. angustus is indeed a single-sex individual, which is sequenced at the gametophyte phase of its life cycle.
Genomic DNA was isolated using the Plant DNAzol reagent for genomic DNA extraction (Life Technologies) according to the manufacturer’s protocols. For whole-genome shotgun sequencing, ten sequencing libraries with insert sizes ranging from 170 bp to 40 kb were generated (Supplementary Table 1). Sequencing libraries were constructed using a library construction kit (Illumina). All libraries were sequenced on the Illumina HiSeq 2000 platform. Raw sequencing reads were trimmed with Trimmomatic (v.0.33)58. Only high-quality reads with a total length of 126,532,381,412 bp were used for further analysis (Supplementary Table 1). For Oxford Nanopore sequencing, we constructed a genomic DNA library using the ONT 1D ligation sequencing kit (SQK-LSK108) according to the manufacturer’s instructions. The sequencing used a single 1D flow cell on a PromethION sequencer (Oxford Nanopore Technologies). A total of 63,614,292,295 bp raw reads were generated, of which 36,070,452,175 bp were retained for further analysis after filtering and trimming (Supplementary Table 3).
Total RNA was extracted using the PureLink Plant RNA reagent (Life Technologies) and further purified using TRIzol reagent (Invitrogen). For transcriptome sequencing (RNA sequencing), libraries with insert sizes ranging from 200 bp to 500 bp were constructed using the mRNA-Seq Prep Kit (Illumina) and then sequenced using the Illumina HiSeq 2000 platform. For small-RNA sequencing, the library was generated from RNA sample using the Truseq Small RNA Preparation kit (Illumina) and sequenced on the Illumina HiSeq 2500 platform.
Decontamination
The GC content versus k-mer frequency distribution pattern of the Illumina raw reads (Supplementary Table 1) after trimming presented two large groups: one group with a low k-mer frequency (<50) and a wide GC content distribution range (median number at 0.7), and the other group with a high k-mer frequency (60–165) and a concentrated GC content distribution range (median number at 0.5) (Supplementary Fig. 2a). The BLASTN results against the NCBI nucleotide database revealed that the former sequences were mainly from a variety of bacteria and the latter were the real genome sequences of A. angustus. We also investigated the k-mer distributions of the raw reads from the other two published hornwort genomic sequences, A. agrestis (accession: ERX714368)59 and Anthoceros punctatus (accession: SRX538621)60, and found a similar distribution pattern as that of A. angustus, containing two groups, one for the contaminant sequences and the other for sequences of the plant itself (Supplementary Fig. 2c,d). Because external bacterial contaminations from the laboratory cause A. angustus to turn yellow and die during culturing, and all three Anthoceros species through axenic cultures still have the same bacterial contamination problems (Supplementary Fig. 2a,c,d), we infer that these bacterial contaminations are from symbiotic bacteria of Anthoceros that might accompany spores hiding in the sterilized sporangium. Furthermore, we performed the DAPI staining analysis61 to investigate the distribution of symbiotic bacteria in A. angustus. The gametophytes were stained by 0.2 mg l−1 DAPI (4′,6-diamidino-2-phenylindole dihydrochloride; Sigma, cat. no. D9564) for five minutes. The stained gametophytes were washed three times, and then observed using confocal microscopy. The bacterial micro-colonies were observed on the outer surface, as well as in the intercellular space of the gametophytes of A. angustus (Supplementary Fig. 3). Based on the GC content versus k-mer frequency distribution pattern of the Illumina raw reads and the result of the DAPI staining, we could imagine that there is a certain amount of bacterial sequences remaining in the genome sequencing data of A. angustus. In order to isolate them, we performed a series of decontamination steps. After generating the k-mer frequency, we chose the high-abundance k-mer depth (60–165) and retained the corresponding reads for further analysis. This treatment yielded filtered reads with a total length of 17,099,027,576 bp (Supplementary Table 2). The distribution pattern of GC content versus k-mer frequency of the A. angustus filtered reads is depicted in Supplementary Fig. 2b, which shows an entire group with a sequencing depth of approximately 150×. Furthermore, we performed error correction for filtered Nanopore reads using decontaminated Illumina reads by Nextdenovo (v.2.0)62, resulting in 9,247,957,448 bp corrected reads (Supplementary Table 3). Through MEGABLAST against the NCBI nucleotide database, we further removed 5,463,972,682 bp prokaryotic sequences or organellar sequences, and finally got 3,783,984,766 clean reads with a sequencing depth of approximately 35× (Supplementary Table 3). A total of approximately 185× coverage was obtained finally.
Genome size estimation
To estimate the genome size of A. angustus, we used clean Illumina reads to calculate the k-mer distribution. According to the Lander–Waterman theory63, the genome size can be determined by dividing the total number of k-mers by the peak value of the k-mer distribution. Because we sequenced the haploid gametophyte of A. angustus, only one peak was found in the k-mer distribution. The total number of k-mers was 14,092,039,150, and the position of the peak was at 132 (Supplementary Fig. 4). The peak was used as the expected k-mer depth and substituted into the formula genome size = total k-mer/expected k-mer depth, and the haploid genome size was estimated to be 106,757,872 bp (Supplementary Fig. 4).
Genome assembly and assessment
The clean Nanopore reads after filtering and decontamination were assembled with wtdbg-1.2.8. After finishing the pre-assembly (148 Mb), iterative polishing was conducted using Pilon (v.1.22)64 in which clean Illumina reads were aligned with the pre-assembled contigs. The pre-assembled contig sequences were performed with the MEGABLAST search against the NCBI nucleotide database to further remove prokaryotic sequences or organellar DNA. A total of approximately 29 Mb of data were removed. Further, we combined the final pre-assembled contig sequences from Nanopore sequencing and clean paired-read data from Illumina sequencing into scaffolds using SSPACE (v.3.0)65 tool (Supplementary Table 4). Genome assembly completeness was assessed using the plantae database of 956 single-copy orthologues using BUSCO (v.3)16 with a BLAST threshold E-value of 1 × 10−5 (Supplementary Table 10).
Transcriptome assembly and mapping
We used Trimmomatic58 to remove adaptors from the raw reads of transcriptome sequences and filter out low-quality reads before assembly. The resulting high-quality reads were de novo assembled and annotated using Trinity (v.2.5.1)66. For genes with more than one transcript, the longest transcript was chosen as the unigene and used to predict open reading frames (ORFs) using TransDecoder (v.5.0.2) (https://github.com/TransDecoder/TransDecoder/wiki). Finally, we obtained 39,044 unigenes, 26,805 of which had predicted ORFs. To extend the validation of genome assembly, the transcriptome was compared to the reference assembly using BLASTN, with an E-value <1 × 10−5. Of the 26,805 transcripts (>200 bp), 97.66% were successfully mapped back to the final assembled genome (Supplementary Table 5).
Repeat prediction
Tandem Repeats Finder (v.4.09)67 was used to search for tandem repeats in the A. angustus genome. Both homology-based and de novo approaches were used to search for TEs. In the homology-based approach, we used RepeatMasker (v.4.1.0)67 and RepeatProteinMask68 with the Repbase69 database of known repeat sequences to search for the TEs in the A. angustus genome. In the de novo approach, we used LTR_FINDER (v.1.0.2)70, PILER (v.1.3.4.)71 and RepeatModeler (v.1.0.3)72 to construct a de novo repeat sequence database for A. angustus and then used RepeatMasker to search for repeats in the genome. All the repeats identified by different methods were combined into the final repeat annotation after removing the redundant repeats. The predicted repeats covered 64.21% of the genome sequence (Supplementary Table 6). The categories of predicted TEs in the A. angustus genome are summarized in Supplementary Table 7.
Genome annotation
To predict protein-coding genes, three approaches were used: (1) de novo gene prediction, (2) homology-based prediction, and (3) RNA-sequencing annotation. For de novo prediction, AUGUSTUS (v.2.5.5)73 and GlimmerHMM (v3.0.1)74 were applied to predict genes. For homology-based prediction, we mapped the protein sequences of five published green plant genomes (Arabidopsis thaliana, Selaginella moellendorffii, P. patens, M. polymorpha and Klebsormidium nitens) onto the A. angustus genome using TBLASTN, with a threshold E-value of 1 × 10−5, and then used GeneWise (v.2.4.1)75 to predict gene structures. The de novo set and five homologue-based results were combined by MAKER (v.1.0)76 to integrate a consensus gene set (Supplementary Table 8). To supplement and improve the gene set, we aligned the RNA-sequencing data to the genome using TopHat (v2.1.1)77, and the alignments were used as input for Cufflinks (v.2.2.1)78 with default parameters. We manually combined the MAKER gene set and ORFs of transcripts to form the final gene set that contains 14,629 genes (Supplementary Table 8).
The A. angustus predicted genes were aligned against the sequences in NCBI non-redundant protein database using BLASTP79 (E-value <1 × 10−5). According to the NCBI taxonomy categories of best BLAST hits, the source of A. angustus genes were classified (Supplementary Fig. 6). Functional annotation of these predicted genes was obtained by aligning the protein sequences of these genes against the sequences in public protein databases using BLASTP79 (E-value <1 × 10−5, identity >30% and coverage >70%, excluding annotations only characterized as hypothetical or predicted protein), including, SwissProt80, TrEMBL80, Pfam81, GO82 and KEGG83 (Supplementary Tables 9 and 32).
Identification of non-coding RNA genes
To obtain a reliable profile of A. angustus miRNAs, we used mapped reads from small-RNA sequencing with reference to the A. angustus draft genome to search against miRNA sequences in A. thaliana, Oryza sativa, S. moellendorffii, P. patens and C. reinhardtii from miRBase (http://www.mirbase.org/) for predicting the known miRNAs. The mapped reads were also used to identify novel miRNAs using miREvo (v.1.2)84 software. The tRNA genes were searched by tRNAscan-SE (v.1.3.1)85. The rRNA genes were predicted by aligning plant rRNA sequences from NCBI (A. thaliana and Anthoceros agrestis) to the A. angustus genome by BLASTN. The snRNA genes were predicted using INFERNAL (v.1.1)86 to search from the Rfam database.
Gene-family identification
To construct the dataset for gene-family clustering, the protein-coding genes from the genomes of A. angustus and 18 other green plants were used, including those of seven angiosperms (A. thaliana, Genlisea aurea, Vitis vinifera, O. sativa, Phalaenopsis equestris, Zostera marina and Amborella trichopoda), one gymnosperm (Picea abies), one lycophyte (S. moellendorffii), two bryophytes (moss P. patens and liverwort M. polymorpha), two charophytes (Chara braunii and K. nitens) and five chlorophytes (Volvox carteri, Chlamydomonas reinhardtii, Ulva mutabilis, Coccomyxa subellipsoidea and Chlorella variabilis) (Supplementary Table 13). We chose the longest transcript to represent each gene and removed mitochondrial and chloroplast genes. After performing an all-against-all BLASTP search with a threshold E-value of 1 × 10−5, identity >30% and coverage >30%, orthogroups or putative gene families or subfamilies were identified using OrthoMCL (v.2.0)87, on the basis of a collection of 397,132 predicted protein-coding genes from the above 19 Viridiplantae genomes. A 5-way comparison of A. angustus, Setaphyta (M. polymorpha and P. patens), Tracheophyta (vascular plants) (A. thaliana, V. vinifera, O. sativa, Z. marina, P. equestris, A. trichopoda, P. abies, G. aurea and S. moellendorffii), Charophyta (C. braunii and K. nitens) and Chlorophyta (V. carteri, C. reinhardtii, U. mutabilis, C. subellipsoidea and C. variabilis) is shown in Fig. 1a. For A. angustus-specific gene families, we conducted GO and KEGG enrichment analyses via an enrichment pipeline (https://sourceforge.net/projects/enrichmentpipeline/).
Phylogenomics
We extracted 85 single-copy gene families shared by 19 Viridiplantae for phylogenomic analysis (Supplementary Note 2.1). The amino acid alignments of each single-copy gene family were aligned by MAFFT (v.7)88, and the nucleotide alignments were generated separately with TranslatorX (v0.9)89 on the basis of the corresponding amino acid translation. The amino acid data, the complete nucleotide data and the first and second codon positions, as well as the third codon positions, were concatenated as super-matrices. These data matrices were used for maximum likelihood phylogenetic analyses by RAxML (v.7.2.3)90 with the GTR + Γ and JTT models for nucleotide and amino acid data, respectively. For each analysis, the bootstrap support was estimated based on 300 pseudoreplicates using a GTR + CAT approximation. To estimate the degree of substitutional saturation for the four concatenated datasets mentioned above (Supplementary Note 2.2), we plotted the uncorrected p-distances against the inferred distances using the method described by Forterre and Philippe91. The level of saturation was estimated by computing the slope of the regression line in the plot; the shallower the slope, the greater the degree of saturation. The maximum composite likelihood method was used to calculate the inferred distances for nucleotide data and Poisson correction was used to calculate the inferred distances for the amino acid data.
To improve the taxon sampling in bryophytes for divergence time estimation, the transcriptome sequences of 22 other bryophytes were downloaded from the 1KP database92 (http://www.onekp.com/public_data.html) and used in subsequent analyses (Supplementary Table 18 and Supplementary Note 2.3). The divergence time was estimated using the MCMCTree program in the PAML package (v.4.7)93 under the nucleotide general time reversible (GTR) substitution model and with the independent rate model as the molecular clock model. The Markov chain Monte Carlo (MCMC) process consists of 500,000 burn-in iterations and 1,500,000 sampling iterations (1 sample per 150 iterations). The same parameters were executed twice to obtain a stable result. We applied nine node constraints in the age estimate (Supplementary Fig. 10). The minimum and maximum constraints for each node are shown in Supplementary Table 19.
Gene-family sizes were inferred from the gene-family profile obtained by the program OrthoMCL. The minimum ancestral gene families were estimated using DOLLOP program included in the PHYLIP package (v.3.695)94 to determine gene-family gain or loss evolutions of gene families. There are 8,141 gene families in the A. angustus genome, 8,944 in M. polymorpha and 9,566 in P. patens, and 9,789 ancestral families in the ancestral bryophyte lineage (Fig. 1b).
K S distribution and co-linearity analysis
All KS distributions were constructed using wgd (v.3.0)95 using default settings. The M. polymorpha and P. patens genome data was acquired from the PLAZA resource96. Pairwise co-linearity analyses within and between A. angustus, M. polymorpha and P. patens were conducted using I-ADHoRe 3.097 with the following parameter settings: gap_size = 30, cluster_gap = 35, q_value = 0.75, prob_cutoff = 0.01, anchor_points = 3, alignment_method = ‘gg2’, level_2_only = ‘false’, table_type = ‘family’ and multiple_hypothesis_correction = ‘FDR’. Within-genome co-linearity analyses were based on the paralogous families inferred with wgd, whereas the between-genome co-linearity analyses were conducted using gene families inferred with OrthoFinder using default settings.
Analysis of TFs
We used the genome-wide TF prediction program iTAK (v.1.7)98 (http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi) with default parameters to preliminarily identify TFs in the above 19 Viridiplantae (Supplementary Tables 13 and 22). The reconstruction of the ancestral state for the individual TF family was performed using Mesquite (v.3.51)99 (http://mesquiteproject.org/), and the most parsimonious assumption was taken.
Phylogenetic analysis of gene families
Generally, HMMER search100 with a domain profile or BLAST search using known protein sequences from other plants as queries was performed to retrieve the sequences from the A. angustus genome (Supplementary Notes 4–6). The results of TF prediction by iTAK98 were used as references. Multiple sequence alignments were performed using the MAFFT88 program (https://mafft.cbrc.jp/alignment/software/). The maximum-likelihood phylogenetic trees were implemented with RAxML-HPC2 on XSEDE101 through the CIPRES Science Gateway (v.3.3) (https://www.phylo.org/), estimating branch support values by bootstrap iterations with 1,000 replicates.
Gene-family expansion identification
To understand gene-family expansion or contraction in A. angustus compared with that in 18 other green plants, the mean gene-family size was calculated for all gene families (excluding orphans and species-specific families). The number of genes per species for each family was transformed into a matrix of z-scores to centre and normalize the data. The first 100 families with the largest gene-family size in A. angustus were selected (Supplementary Fig. 55). The clustering and visualization were performed using Genesis (v.3.0)102. The functional annotation of each family was predicted on the basis of sequence similarity to entries in the Pfam protein domain database, where more than 30% of proteins in the family share the same protein domain. Transposon-derived gene families were removed because the distribution of such families is likely to be a consequence of the gene models derived from a repeat-masked genome sequence and therefore may be artefactual103.
Tandem duplication definition
Genes were defined as tandemly arrayed genes if they belonged to the same family, were located within 100 kb each other, and were separated by zero, one or fewer, five or fewer, or ten or fewer non-homologous intervening ‘spacer’ genes104. Therefore, the four sets of tandem gene definitions were analysed.
HGT event identification
In this study, we used two different strategies to identify candidates for A. angustus-specific and bryophyte-specific HGTs. For A. angustus-specific HGTs, we submitted 14,629 predicted coding genes of A. angustus to a BLASTP search against the NCBI protein database (E-value cutoff of 1 × 10−5) (Supplementary Note 7.1). The proteins with the best BLAST hits in bacterial or fungal sequences were extracted. After sequences without support of transcript evidence were excluded, a series of parameters were used to filter the candidates (Supplementary Note 7.1). For the bryophyte-specific HGT, we extracted gene families that are common to at least two of the three members of bryophytes (moss P. patens, liverwort M. polymorpha and hornwort A. angustus). To preliminarily determine whether these clusters are HGT candidates, we submitted the corresponding A. angustus members of each cluster to the NCBI protein database for BLASTP search and checked the taxonomy report of the top 1,000 BLAST hits (Supplementary Note 7.2). The homologues of published HGTs in P. patens51 and M. polymorpha15 were also investigated in the A. angustus genome. All candidate HGTs were subjected to phylogenetic analysis for verification. Synonymous codon-usage order values and GC contents of HGT and non-HGT genes were calculated by CodonO105.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The A. angustus genome project has been deposited at the NCBI under the BioProject number PRJNA543716. The genome sequencing data were deposited in the Sequence Read Archive database under the accession number SRR9696346. The A. angustus transcriptome project has been deposited at the NCBI under BioProject PRJNA543724. The transcriptome sequencing data were deposited in the Sequence Read Archive database under the accession number SRR9662965. The assembled genome sequences, gene models and miRNA data are available via DRYAD (https://doi.org/10.5061/dryad.msbcc2ftv). All data that support the findings of this study are also available from the corresponding authors upon request.
References
Puttick, M. N. et al. The interrelationships of land plants and the nature of the ancestral embryophyte. Curr. Biol. 28, 733–745 (2018).
Goffinet, B. & Buck, W. R. in The Evolution of Plant Form (eds Ambrose, B. & Purruganan, M.) 51–90 (Wiley–Blackwell, 2013).
von Konrat, M., Shaw, A. J. & Renzaglia, K. S. A special issue of Phytotaxa dedicated to bryophytes: the closest living relatives of early land plants. Phytotaxa 9, 5–10 (2010).
Christenhusz, M. J. M. & Byng, J. W. The number of known plants species in the world and its annual increase. Phytotaxa 261, 201–217 (2016).
Qiu, Y. L. et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc. Natl Acad. Sci. USA 103, 15511–15516 (2006).
Wickett, N. J. et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl Acad. Sci. USA 111, E4859–E4868 (2014).
Cox, C. J., Li, B., Foster, P. G., Embley, T. M. & Civan, P. Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions. Syst. Biol. 63, 272–279 (2014).
Liu, Y., Cox, C. J., Wang, W. & Goffinet, B. Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias. Syst. Biol. 63, 862–878 (2014).
Villarreal, J. C. & Renzaglia, K. S. The hornworts: important advancements in early land plant evolution. J. Bryol. 37, 157–170 (2015).
Villarreal, J. C. & Renner, S. S. Hornwort pyrenoids, carbon-concentrating structures, evolved and were lost at least five times during the last 100 million years. Proc. Natl Acad. Sci. USA 109, 18873–18878 (2012).
Renzaglia, K. S., Villarreal, J. C. & Duff, R. J. in Bryophyte Biology (eds Goffinet, B. & Shaw, J.) 139–171 (Cambridge Univ. Press, 2009).
Adams, D. G. & Duggan, P. S. Cyanobacteria–bryophyte symbioses. J. Exp. Bot. 59, 1047–1058 (2008).
Desirὸ, A., Duckett, J. G., Pressel, S., Villarreal, J. C. & Bidartondo, M. I. Fungal symbioses in hornworts: a chequered history. Proc. R. Soc. B 280, 1759 (2013).
Rensing, S. A. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008).
Bowman, J. L. et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304 (2017).
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Axtell, M. J. & Bowman, J. L. Evolution of plant microRNAs and their targets. Trends Plant Sci. 13, 343–349 (2008).
Archangelsky, S. & Villar de Seone, L. Estudios palinógicos de la formación Baqueró (Cretácico), provincia de Santa Cruz, Argentina. Ameghiniana 35, 7–19 (1996).
Van de Peer, Y., Mizrachi, E. & Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017).
Lang, D. et al. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 93, 515–533 (2018).
Catarino, B., Hetherington, A. J., Emms, D. M., Kelly, S. & Dolan, L. The stepwise increase in the number of transcription factor families in the Precambrian predated the diversification of plants on land. Mol. Biol. Evol. 33, 2815–2819 (2016).
Cheng, F. et al. Gene retention, fractionation and subgenome differences in polyploid plants. Nat. Plants 4, 258–268 (2018).
Lang, D. et al. Genome-wide phylogenetic comparative analysis of plant transcriptional regulation: a timeline of loss, gain, expansion, and correlation with complexity. Genome Biol. Evol. 2, 488–503 (2010).
Sakakibara, K. Technological innovations give rise to a new era of plant evolutionary developmental biology. Adv. Bot. Res. 78, 3–35 (2016).
Szövényi, P., Waller, M. & Kirbis, A. Evolution of the plant body plan. Curr. Top. Dev. Biol. 131, 1–34 (2019).
Floyd, S. K. & Bowman, J. L. The ancestral developmental tool kit of land plants. Int. J. Plant Sci. 168, 1–35 (2007).
Hori, K. et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5, 3978 (2014).
Nishiyama, T. et al. The Chara genome: secondary complexity and implications for plant terrestrialization. Cell 174, 448–464 (2018).
Wodniok, S. et al. Origin of land plants: do conjugating green algae hold the key? BMC Evol. Biol. 11, 104 (2011).
Ishizaki, K. Evolution of land plants: insights from molecular studies on basal lineages. Biosci. Biotechnol. Biochem. 81, 73–80 (2017).
Rensing, S. A. Great moments in evolution: the conquest of land by plants. Curr. Opin. Plant Biol. 42, 49–54 (2018).
Braybrook, S. A. & Harada, J. J. LECs go crazy in embryo development. Trends Plant Sci. 13, 624–630 (2008).
Takezawa, D., Komatsu, K. & Sakata, Y. ABA in bryophytes: how a universal growth regulator in life became a plant hormone? J. Plant Res. 124, 437–453 (2011).
Proust, H. et al. RSL class I genes controlled the development of epidermal structures in the common ancestor of land plants. Curr. Biol. 26, 93–99 (2016).
Pires, N. D. et al. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl Acad. Sci. USA 110, 9571–9576 (2013).
Sakakibara, K. et al. KNOX2 genes regulate the haploid-to-diploid morphological transition in land plants. Science 339, 1067–1070 (2013).
Coudert, Y., Novák, O. & Harrison, C. J. A KNOX-cytokinin regulatory module predates the origin of indeterminate vascular plants. Curr. Biol. 29, 2743–2750 (2019).
Mutte, S. K. et al. Origin and evolution of the nuclear auxin response system. eLife 7, e33399 (2018).
Cenci, A. & Rouard, M. Evolutionary analyses of GRAS transcription factors in angiosperms. Front. Plant Sci. 8, 273 (2017).
Kaplan-Levy, R. N., Brewer, P. B., Quon, T. & Smyth, D. R. The trihelix family of transcription factors—light, stress and development. Trends Plant Sci. 17, 163–171 (2012).
Fujii, S. & Small, I. The evolution of RNA editing and pentatricopeptide repeat genes. N. Phytol. 191, 37–47 (2011).
Cheng, S. et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547 (2016).
Rüdinger, M., Polsakiewicz, M. & Knoop, V. Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat proteins in jungermanniid but not in marchantiid liverworts. Mol. Biol. Evol. 25, 1405–1414 (2008).
Dunwell, J. M., Khuri, S. & Gane, P. J. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64, 153–179 (2000).
Nakata, M. et al. Germin-like protein gene family of a moss, Physcomitrella patens, phylogenetically falls into two characteristic new clades. Plant Mol. Biol. 56, 381–395 (2004).
Pollastri, S. & Tattini, M. Flavonols: old compounds for old roles. Ann. Bot. 108, 1225–1233 (2011).
Sakata, Y., Komatsu, K. & Takezawa, D. in Progress in Botany (ed. Lüttge, U.) 57–96 (Springer-Verlag, 2014).
Hanson, D. T., Renzaglia, K. & Villareal, J. C. in Photosynthesis of Bryophytes and Early Land Plants (eds Hanson, D. T. & Rice, S. K.) 95–111 (Springer, 2014).
Meyer, M. & Griffiths, H. Origins and diversity of eukaryotic CO2-concentrating mechanisms: lessons for the future. J. Exp. Bot. 64, 769–786 (2013).
Mackinder, L. C. M. A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171, 133–147 (2017).
Yue, J., Hu, X., Sun, H., Yang, Y. & Huang, J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat. Commun. 3, 1152 (2012).
Foflonker, F. et al. Genome of the halotolerant green alga Picochlorum sp. reveals strategies for thriving under fluctuating environmental conditions. Environ. Microbiol. 17, 412–426 (2015).
Hasanuzzaman, M. et al. Coordinated actions of glyoxalase and antioxidant defense systems in conferring abiotic stress tolerance in plants. Int. J. Mol. Sci. 18, 200 (2017).
Finnegan, E. J. & Kovac, K. A. Plant DNA methyltransferases. Plant Mol. Biol. 43, 189–201 (2000).
Jia, Q. et al. Microbial-type terpene synthase genes occur widely in nonseed land plants, but not in seed plants. Proc. Natl Acad. Sci. USA 113, 12328–12333 (2016).
Duckett, J. G. et al. In vitro cultivation of bryophytes: a review of practicalities, problems, progress and promise. J. Bryol. 26, 3–20 (2004).
Kugita, M. et al. The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: insight into the earliest land plants. Nucleic Acids Res. 31, 716–721 (2003).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Szövényi, P. et al. Establishment of Anthoceros agrestis as a model species for studying the biology of hornworts. BMC Plant Biol. 15, 98 (2015).
Li, F. et al. Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proc. Natl Acad. Sci. USA 111, 6672–6677 (2014).
Mergaert, P. et al. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proc. Natl Acad. Sci. USA 103, 5230–5235 (2006).
Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Arratia, R., Lander, E. S., Tavaré, S. & Waterman, M. S. Genomic mapping by anchoring random clones: a mathematical analysis. Genomics 11, 806–827 (1991).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014).
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D. & Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2011).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics 25, 4.10.1–4.10.14 (2009).
Jurka, J. et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007).
Edgar, R. C. & Myers, E. W. PILER: identification and classification of genomic repeats. Bioinformatics 21(Suppl. 1), i152–i158 (2005).
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21(Suppl. 1), i351–i358 (2005).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439 (2006).
Majoros, W. H., Pertea, M. & Salzberg, S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004).
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res. 14, 988–995 (2004).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinf. 12, 491 (2011).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinf. 10, 421 (2009).
Boeckmann, B. et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370 (2003).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Wen, M., Shen, Y., Shi, S. & Tang, T. miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinf. 13, 140 (2012).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013).
Li, L., Stoeckert, C. J. & Roos, D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003).
Katoh, K., Kuma, K. I., Toh, H. & Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005).
Abascal, F., Zardoya, R. & Telford, M. J. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38, W7–W13 (2010).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Forterre, P. & Philippe, H. Where is the root or the universal tree of life? Bioessays 21, 871–879 (1999).
Matasci, N. et al. Data access for the 1,000 Plants (1KP) project. Gigascience 3, 17 (2014).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Felsenstein, J. PHYLIP: phylogenetic inference program v.3.6 (Univ. of Washington, 2005).
Zwaenepoel, A. & Van de Peer, Y. wgd—simple command line tools for the analysis of ancient whole genome duplications. Bioinformatics 35, 2153–2155 (2018).
Van Bel, M. et al. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46, D1190–D1196 (2018).
Proost, S. et al. i-ADHoRe 3.0―fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 40, e11 (2012).
Zheng, Y. et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670 (2016).
Maddison, W. P. & Maddison, D. R. Mesquite: a modular system for evolutionary analysis v.2.75 (Mesquite Project, 2011).
Madera, M. & Gough, J. A comparison of profile hidden Markov model procedures for remote homology detection. Nucleic Acids Res. 30, 4321–4328 (2002).
Stamatakis, A., Hoover, P. & Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771 (2008).
Sturn, A., Quackenbush, J. & Trajanoski, Z. Genesis: cluster analysis of microarray data. Bioinformatics 18, 207–208 (2002).
Martens, C., Vandepoele, K. & Van de Peer, Y. Whole-genome analysis reveals molecular innovations and evolutionary transitions in chromalveolate species. Proc. Natl Acad. Sci. USA 105, 3427–3432 (2008).
Hanada, K. et al. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148, 993–1003 (2008).
Angellotti, M. C., Bhuiyan, S. B., Chen, G., Wan, X. & Wan, X. CodonO: codon usage bias analysis within and across genomes. Nucleic Acids Res. 35, W132–W136 (2007).
Acknowledgements
We thank P. R. Crane, S. Ge, D.-Y. Hong, J.-L. Huang, J.-J. Qin, Y.-L. Qiu, J.-C. Villarreal, T. Wan and X.-Q. Wang for useful advice and discussions and L. Zhang for providing plant pictures. We dedicate the paper to Yang Zhong in memory of his support and valuable suggestions on this project. This work was supported by Sino–Africa Joint Research Center, Chinese Academy of Sciences, CAS International Research and Education Development Program (SAJC201613), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31000000 and XDA19050103), National Natural Science Foundation of China (NNSF 31590822), Shenzhen Fairy Lake Botanical Garden, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, and Key Laboratory of National Forestry and Grassland Administration for Orchid Conservation and Utilization at College of Landscape Architecture, Fujian Agriculture and Forestry University. Y.V.d.P. acknowledges support from the European Union Seventh Framework Programme (FP7/2007-2013) under European Research Council Advanced Grant Agreement 322739–DOUBLEUP.
Author information
Authors and Affiliations
Contributions
Z.-D.C., Z.-J.L., Y.V.d.P. and S.-Z.Z. conceived the paper; Z.-D.C., Z.-J.L. and S.-Z.Z. managed the project; J.Z., X.-X.F., Y.L., Z.-J.L., A.Z., Y.V.d.P. and Z.-D.C. wrote the manuscript; R.-Q.L., J.-F.Y., Y.-Y.L., Q.-H.W., S.-Z.Z. and M.-Z.W. collected and cultured the plant material; R.-Q.L. and M.-H.L. sequenced and processed the raw data; X.Z. and Z.-W.W. assembled and annotated the genome; Y.L. and J.Z. performed phylogenetic analysis; J.Z. and X.-X.F. analysed gene families; X.-X.F. and J.Z. identified HGT; A.Z. and Y.V.d.P. conducted WGD analysis; Y.-L. Guan. conducted DAPI staining analysis; J.-Y.X. conducted codon-usage bias analysis; M.-H.L., G.-Q.Z. and J.-Y.W. conducted transcriptome sequencing and analysis; S.-S.D. and Y.L. conducted the RNA-editing-site analysis in organellar genomes; H.M., Q.-F.W., B.G., Y.J., Y.-N.J., Y.-L.Guo, H.-Z.K., A.-M.L. and H.-M.Y. contributed substantially to revisions. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Peer review information Nature Plants thanks Burkhard Becker and the other, anonymous, reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–7, Figs. 1–87 and Tables 1–20.
Supplementary Tables
Supplementary Tables 21–32.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, J., Fu, XX., Li, RQ. et al. The hornwort genome and early land plant evolution. Nat. Plants 6, 107–118 (2020). https://doi.org/10.1038/s41477-019-0588-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0588-4
This article is cited by
-
Comparative analysis of SPL transcription factors from streptophyte algae and embryophytes reveals evolutionary trajectories of SPL family in streptophytes
Scientific Reports (2024)
-
A mysterious cloak: the peptidoglycan layer of algal and plant plastids
Protoplasma (2024)
-
Theme and variations
Nature Plants (2023)
-
The first homosporous lycophyte genome revealed the association between the recent dynamic accumulation of LTR-RTs and genome size variation
Plant Molecular Biology (2023)
-
Bryophytes as Modern Model Plants: An Overview of Their Development, Contributions, and Future Prospects
Journal of Plant Growth Regulation (2023)