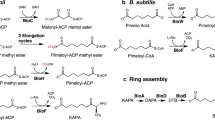
Abstract
In biotin biosynthesis, the conversion of pimeloyl intermediates to biotin is catalyzed by a universal set of four enzymes: BioF, BioA, BioD and BioB. We found that the gene homologous to bioA, the product of which is involved in the conversion of 8-amino-7-oxononanoate (AON) to 7,8-diaminononanoate (DAN), is missing in the genome of the cyanobacterium Synechocystis sp. PCC 6803. We provide structural and biochemical evidence showing that a novel dehydrogenase, BioU, is involved in biotin biosynthesis and functionally replaces BioA. This enzyme catalyzes three reactions: formation of covalent linkage with AON to yield a BioU-DAN conjugate at the ε-amino group of Lys124 of BioU using NAD(P)H, carboxylation of the conjugate to form BioU-DAN-carbamic acid, and release of DAN-carbamic acid using NAD(P)+. In this biosynthetic pathway, BioU is a suicide enzyme that loses the Lys124 amino group after a single round of reaction.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
References
Wood, H. G. & Barden, R. E. Biotin enzymes. Annu. Rev. Biochem. 46, 385–413 (1977).
Hansen, D. E. & Knowles, J. R. N-carboxybiotin formation by pyruvate carboxylase: the stereochemical consequence at phosphorus. J. Am. Chem. Soc. 107, 8304–8305 (1985).
Zempleni, J., Wijeratne, S. S. & Hassan, Y. I. Biotin. Biofactors 35, 36–46 (2009).
Sherwood, W. G., Saunders, M., Robinson, B. H., Brewster, T. & Gravel, R. A. Lactic acidosis in biotin-responsive multiple carboxylase deficiency caused by holocarboxylase synthetase deficiency of early and late onset. J. Pediatr. 101, 546–550 (1982).
Seymons, K., De Moor, A., De Raeve, H. & Lambert, J. Dermatologic signs of biotin deficiency leading to the diagnosis of multiple carboxylase deficiency. Pediatr. Dermatol. 21, 231–235 (2004).
Lin, S. & Cronan, J. E. Closing in on complete pathways of biotin biosynthesis. Mol. Biosyst. 7, 1811–1821 (2011).
Stoner, G. L. & Eisenberg, M. A. Purification and properties of 7,8-diaminopelargonic acid aminotransferase. J. Biol. Chem. 250, 4029–4036 (1975).
Van Arsdell, S. W. et al. Removing a bottleneck in the Bacillus subtilis biotin pathway: BioA utilizes lysine rather than S-adenosylmethionine as the amino donor in the KAPA-to-DAPA reaction. Biotechnol. Bioeng. 91, 75–83 (2005).
Wierenga, R. K., De Maeyer, M. C. H. & Hol, W. G. J. Interaction of pyrophosphate moieties with alpha-helixes in dinucleotide binding proteins. Biochemistry 24, 1346–1357 (1985).
Hudson, R. C. & Daniel, R. M. l-Glutamate dehydrogenases: distribution, properties and mechanism. Comp. Biochem. Physiol. B 106, 767–792 (1993).
Tang, G., Miron, D., Zhu-Shimoni, J. X. & Galili, G. Regulation of lysine catabolism through lysine-ketoglutarate reductase and saccharopine dehydrogenase in Arabidopsis. Plant Cell 9, 1305–1316 (1997).
Zhu, X., Tang, G. & Galili, G. The activity of the Arabidopsis bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase enzyme of lysine catabolism is regulated by functional interaction between its two enzyme domains. J. Biol. Chem. 277, 49655–49661 (2002).
Sheng, X., Gao, J., Liu, Y. & Liu, C. Theoretical study on the proton shuttle mechanism of saccharopine dehydrogenase. J. Mol. Graph. Model. 44, 17–25 (2013).
Vashishtha, A. K., West, A. H. & Cook, P. F. Probing the chemical mechanism of saccharopine reductase from Saccharomyces cerevisiae using site-directed mutagenesis. Arch. Biochem. Biophys. 584, 98–106 (2015).
Huang, W. et al. Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate. Biochemistry 34, 10985–10995 (1995).
Yang, G., Sandalova, T., Lohman, K., Lindqvist, Y. & Rendina, A. R. Active site mutants of Escherichia coli dethiobiotin synthetase: effects of mutations on enzyme catalytic and structural properties. Biochemistry 36, 4751–4760 (1997).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Teo, I., Sedgwick, B., Kilpatrick, M. W., McCarthy, T. V. & Lindahl, T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell 45, 315–324 (1986).
Chatterjee, A. et al. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 478, 542–546 (2011).
Lai, R. Y. et al. Thiamin pyrimidine biosynthesis in Candida albicans: a remarkable reaction between histidine and pyridoxal phosphate. J. Am. Chem. Soc. 134, 9157–9159 (2012).
Broquist, H. P. & Snell, E. E. Biotin and bacterial growth. 1. Relation to aspartate, oleate and carbon dioxide. J. Biol. Chem. 188, 431–444 (1951).
Cronan, J. E. Expression of the biotin biosynthetic operon of Escherichia coli is regulated by the rate of protein biotination. J. Biol. Chem. 263, 10332–10336 (1988).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 1989).
Rippka, R. Isolation and purification of cyanobacteria. Methods Enzymol. 167, 3–27 (1988).
Ng, A. H., Berla, B. M. & Pakrasi, H. B. Fine-tuning of photoautotrophic protein production by combining promoters and neutral sites in the cyanobacterium Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 81, 6857–6863 (2015).
Bordat, A., Houvenaghel, M. C. & German-Retana, S. Gibson assembly: an easy way to clone potyviral full-length infectious cDNA clones expressing an ectopic VPg. Virol. J. 12, 89 (2015).
Jez, J. M., Ferrer, J. L., Bowman, M. E., Dixon, R. A. & Noel, J. P. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39, 890–902 (2000).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Pannu, N. S. et al. Recent advances in the CRANK software suite for experimental phasing. Acta Crystallogr. D 67, 331–337 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Laskowski, R. A., Macarthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Acknowledgements
This work was supported in part by JSPS KAKENHI (grant no. 17H06168 to M.N.) and the Research Committee of B group Vitamins (to M.N.). We are grateful to the staff at Photon Factory for their assistance with data collection, which was approved by the Photon Factory Program Advisory Committee (proposals nos. 2015G546, 2017G574 and 2019G548).
Author information
Authors and Affiliations
Contributions
K.S. and K.O performed cloning of bioU genes, biochemical analysis of BioU, crystallographic analysis of BioU and X-ray data correction. K.S. also wrote the manuscript. T.S. performed genome mining for bioU. I.K. and K.T. performed genetics analysis of bioU in Synechocystis sp. PCC 6803. H.W. and N.M. synthesized 7. K.M. analyzed the reaction intermediate. T.T. performed crystallographic analysis and structure refinements. T.K. and M.N. designed the project, analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Figs. 1–14 and Note.
Rights and permissions
About this article
Cite this article
Sakaki, K., Ohishi, K., Shimizu, T. et al. A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria. Nat Chem Biol 16, 415–422 (2020). https://doi.org/10.1038/s41589-019-0461-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0461-9
This article is cited by
-
Advances in biotin biosynthesis and biotechnological production in microorganisms
World Journal of Microbiology and Biotechnology (2024)
-
An outlook on suicide enzyme inhibition and drug design
Journal of the Iranian Chemical Society (2022)