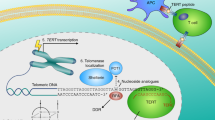
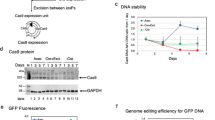
Abstract
The transcription factor NF-κB is an attractive target for cancer therapy due to its over-activation in all tumours; however, NF-κB inhibitors developed in the past decades rarely became drugs due to undesirable side effects. In this study, we developed a gene therapy strategy named NF-κB-activated gene expression (Nage), which could induce the death of cancer cells in vitro and in vivo by utilising NF-κB over-activity in cancer cells, but had no effects on normal cells. Nage was consisted of an NF-κB-specific promoter formed by fusing an NF-κB decoy sequence with a minimal promoter, which could be bound by the intracellular over-activated NF-κB and thus activated the expression of downstream effector genes in an NF-κB-specific manner. In this study, we first confirmed the cancer cell-specific over-activation of NF-κB and then tested the cancer cell specificity of the Nage vector by expressing the reporter gene ZsGreen in various in vitro cultivated cells. We next demonstrated that the Nage vector could be used to express CRISPR/Cas9 protein only in cancer cells. The Cas9 protein was then guided by a sgRNA targeting telomeric DNA and induced cancer cell death. The Nage vector expressing Cas9/sgRNA could be packaged into adeno-associated virus (AAV) and intravenously injected to inhibit tumour growth in mice without visible side effects and toxicity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–16.
Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-κB signaling pathways. Nat Immunol. 2011;12:695–708.
Hatada EN, Krappmann D, Scheidereit C. NF-κB and the innate immune response. Curr Opin Immunol. 2000;12:52–8.
Hayden MS, West AP, Ghosh S. NF-κB and the immune response. Oncogene. 2006;25:6758–80.
Oeckinghaus A, Ghosh S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1:a000034.
Wan F, Lenardo MJ. Specification of DNA binding activity of NF-κB proteins. Cold Spring Harb Perspect Biol. 2009;1:a000067.
Smale ST. Hierarchies of NF-κB target-gene regulation. Nat Immunol. 2011;12:689–94.
Pahl HL. Activators and target genes of Rel/NF-κB transcription factors. Oncogene. 1999;18:6853–66.
Ahn KS, Aggarwal BB. Transcription factor NF-κB—A sensor for smoke and stress signals. Nat Prod Mol Ther. 2005;1056:218–33.
Karin M. NF-κB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1:a000141.
Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat Immunol. 2011;12:715–23.
DiDonato JA, Mercurio F, Karin M. NF-κB and the link between inflammation and cancer. Immunol Rev. 2012;246:379–400.
Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86.
Basseres DS, Baldwin AS. Nuclear factor-κB and inhibitor of kappa B kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817–30.
Barkett M, Gilmore TD. Control of apoptosis by Rel/NF-κB transcription factors. Oncogene. 1999;18:6910–24.
Biswas DK, Cruz AP, Gansberger E, Pardee AB. Epidermal growth factor-induced nuclear factor κB activation: a major pathway of cell-cycle progression in estrogen-receptor negative breast cancer cells. Proc Natl Acad Sci USA. 2000;97:8542–7.
Sen R. Control of B lymphocyte apoptosis by the transcription factor NF-κB. Immunity. 2006;25:871–83.
Ainbinder E, Revach M, Wolstein O, Moshonov S, Diamant N, Dikstein R. Mechanism of rapid transcriptional induction of tumor necrosis factor alpha-responsive genes by NF-κB. Mol Cell Biol. 2002;22:6354–62.
Lane DP, Midgley CA, Hupp TR, Lu X, Vojtesek B, Picksley SM. On the regulation of the p53 tumour suppressor, and its role in the cellular response to DNA damage. Philos Trans R Soc Lond B Biol Sci. 1995;347:83–7.
Su S, Chen J, Yao H, Liu J, Yu S, Lao L, et al. CD10+GPR77+ cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell. 2018;172:841–56.e16.
Nguyen DP, Li JY, Yadav SS, Tewari AK. Recent insights into NF-κB signalling pathways and the link between inflammation and prostate cancer. BJU Int. 2014;114:168–76.
Grinberg-Bleyer Y, Ghosh S. A novel link between inflammation and cancer. Cancer Cell. 2016;30:829–30.
Karin M, Bonnizi G, Cao Y, Delhase M, Greten F, Hu YL, et al. NF-κB: a factor that provides a link between stress, inflammation and cancer. Eur J Cancer. 2002;38:S116.
Karin M, Yamamoto Y, Wang QM. The IKK NF-κB system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26.
Gupta SC, Sundaram C, Reuter S, Aggarwal BB. Inhibiting NF-κB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta. 2010;1799:775–87.
Pikarsky E, Ben-Neriah Y. NF-κB inhibition: a double-edged sword in cancer? Eur J Cancer. 2006;42:779–84.
Xiao G, Rabson AB, Young W, Qing G, Qu Z. Alternative pathways of NF-κB activation: a double-edged sword in health and disease. Cytokine Growth Factor Rev. 2006;17:281–93.
Tornatore L, Sandomenico A, Raimondo D, Low C, Rocci A, Tralau-Stewart C, et al. Cancer-selective targeting of the NF-κB survival pathway with GADD45β/MKK7 inhibitors. Cancer Cell. 2014;26:938.
Rega C, Russo R, Foca A, Sandomenico A, Iaccarino E, Raimondo D, et al. Probing the interaction interface of the GADD45beta/MKK7 and MKK7/DTP3 complexes by chemical cross-linking mass spectrometry. Int J Biol Macromol. 2018;114:114–23.
Wang D, Tang H, Xu X, Dai W, Wu J, Wang J. Control the intracellular NF-κB activity by a sensor consisting of miRNA and decoy. Int J Biochem Cell Biol. 2018;95:43–52.
Wang JK, Li TX, Bai YF, Lu ZH. Evaluating the binding affinities of NF-κB p50 homodimer to the wild-type and single-nucleotide mutant Ig-κB sites by the unimolecular dsDNA microarray. Anal Biochem. 2003;316:192–201.
Zhou F, Xing Y, Xu X, Yang Y, Zhang J, Ma Z, et al. NBPF is a potential DNA-binding transcription factor that is directly regulated by NF-κB. Int J Biochem Cell Biol. 2013;45:2479–90.
Gu G, Wang T, Yang Y, Xu X, Wang J. An improved SELEX-Seq strategy for characterizing DNA-binding specificity of transcription factor: NF-κB as an example. PLoS ONE. 2013;8:e76109.
Xing Y, Zhou F, Wang J. Subset of genes targeted by transcription factor NF-κB in TNFα-stimulated human HeLa cells. Funct Integr Genom. 2013;13:143–54.
Zhou F, Wang W, Xing YJ, Wang TT, Xu XH, Wang JK. NF-κB target microRNAs and their target genes in TNFα-stimulated HeLa Cells. Biochim Biophys Acta. 2014;1839:344–54.
Zhou F, Xu X, Wang D, Wu J, Wang J. Identification of novel NF-κB transcriptional targets in TNFα-treated HeLa and HepG2 cells. Cell Biol Int. 2017;41:555–69.
Dai W, Wu J, Zhang S, Shi B, Xu X, Wang D, et al. Genes directly regulated by NF-κB in human hepatocellular carcinoma HepG2. Int J Biochem Cell Biol. 2017;89:157–70.
Wang D, Dai W, Wu J, Wang J. Improving transcriptional activity of human cytomegalovirus major immediate-early promoter by mutating NF-κB binding sites. Protein Expr Purif. 2018;142:16–24.
Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47.
Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–91.
Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–73.
Moon IK, Jarstfer MB. The human telomere and its relationship to human disease, therapy, and tissue engineering. Front Biosci. 2007;12:4595–620.
Cairney CJ, Keith WN. Telomerase redefined: integrated regulation of hTR and hTERT for telomere maintenance and telomerase activity. Biochimie. 2008;90:13–23.
Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, et al. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–75.
Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–8.
Chen W. Accelerating cancer evolution: a dark side of SIRT1 in genome maintenance. Oncotarget. 2012;3:363–4.
Counter CM, Botelho FM, Wang P, Harley CB, Bacchetti S. Stabilization of short telomeres and telomerase activity accompany immortalization of Epstein-Barr virus-transformed human B lymphocytes. J Virol. 1994;68:3410–4.
Aschacher T, Wolf B, Enzmann F, Kienzl P, Messner B, Sampl S, et al. LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines. Oncogene. 2016;35:94–104.
Deng Y, Chang S. Role of telomeres and telomerase in genomic instability, senescence and cancer. Lab Investig. 2007;87:1071–6.
Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18:173–9.
Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Semin Cancer Biol. 2011;21:349–53.
Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–9.
Jafri MA, Ansari SA, Alqahtani MH, Shay JW. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8:69.
Mosoyan G, Kraus T, Ye F, Eng K, Crispino JD, Hoffman R, et al. Imetelstat, a telomerase inhibitor, differentially affects normal and malignant megakaryopoiesis. Leukemia. 2017;31:2458–67.
Tefferi A, Lasho TL, Begna KH, Patnaik MM, Zblewski DL, Finke CM, et al. A pilot study of the telomerase inhibitor imetelstat for myelofibrosis. N Engl J Med. 2015;373:908–19.
Zeng X, Hernandez-Sanchez W, Xu M, Whited TL, Baus D, Zhang J, et al. Administration of a nucleoside analog promotes cancer cell death in a telomerase-dependent manner. Cell Rep. 2018;23:3031–41.
Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70.
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–7.
Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096.
Samanta MK, Dey A, Gayen S. CRISPR/Cas9: an advanced tool for editing plant genomes. Transgenic Res. 2016;25:561–73.
Hermonat PL, Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc Natl Acad Sci USA. 1984;81:6466–70.
George LA. Hemophilia gene therapy comes of age. Blood Adv. 2017;1:2591–9.
Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med. 2017;377:1713–22.
Valdmanis PN, Kay MA. Future of rAAV gene therapy: platform for RNAi, gene editing, and beyond. Hum Gene Ther. 2017;28:361–72.
Shen S, Sanchez ME, Blomenkamp K, Corcoran EM, Marco E, Yudkoff CJ, et al. Amelioration of Alpha-1 antitrypsin deficiency diseases with genome editing in transgenic mice. Hum Gene Ther. 2018;29:861–73.
Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74.
Louis N, Evelegh C, Graham FL. Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology. 1997;233:423–9.
Funding
This work was supported by the National Natural Science Foundation of China (61971122) and the National Key Research and Development Programme of China (2017YFA0205502).
Author information
Authors and Affiliations
Contributions
JKW conceived the study and designed experiments. WD designed and performed main experiments. JW and DYW prepared reagents and performed partial experiments. JKW and WD wrote the paper with support from all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Dai, W., Wu, J., Wang, D. et al. Cancer gene therapy by NF-κB-activated cancer cell-specific expression of CRISPR/Cas9 targeting telomeres. Gene Ther 27, 266–280 (2020). https://doi.org/10.1038/s41434-020-0128-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-020-0128-x
This article is cited by
-
Sex-dependent associations between MAP3K1 gene polymorphisms and soy products with the gastric cancer risk in Korea: a case-control study
BMC Gastroenterology (2022)
-
Ferroptosis assassinates tumor
Journal of Nanobiotechnology (2022)
-
Gene interfered-ferroptosis therapy for cancers
Nature Communications (2021)