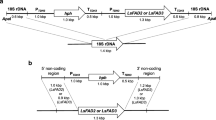
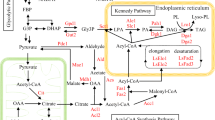
Abstract
The oleaginous yeast Lipomyces starkeyi is a potential cost-effective source for the production of microbial lipids. Fatty acid elongases have vital roles in the syntheses of long-chain fatty acids. In this study, two genes encoding fatty acid elongases of L. starkeyi, LsELO1, and LsELO2 were identified and characterized. Heterologous expression of these genes in Saccharomyces cerevisiae revealed that LsElo1 is involved in the production of saturated long-chain fatty acids with 24 carbon atoms (C24:0) and that LsElo2 is involved in the conversion of C16 fatty acids to C18 fatty acids. In addition, both LsElo1 and LsElo2 were able to elongate polyunsaturated fatty acids. LsElo1 elongated linoleic acid (C18:2) to eicosadienoic acid (C20:2), and LsElo2 elongated α-linolenic acid (C18:3) to eicosatrienoic acid (C20:3). Overexpression of LsElo2 in L. starkeyi caused a reduction in C16 fatty acids, such as palmitic and palmitoleic acids, and an accumulation of C18 fatty acids such as oleic and linoleic acids. Our findings have the potential to contribute to the remodeling of fatty acid composition and the production of polyunsaturated long-chain fatty acids in oleaginous yeasts.


Similar content being viewed by others
References
Ageitos JM, Vallejo JA, Veiga-Crespo P, Villa TG (2011) Oily yeasts as oleaginous cell factories. Appl Microbiol Biotechnol 90:1219–1227
Ballweg and Emst (2017) Control of membrane fluidity: the OLE pathway in focus. 398:215–228
Chellappa R, Kandasamy P, Oh CS, Jiang Y, Vemula M, Martin CE (2001) The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J Biol Chem 276:43548–43556
Das T, Thurmond JM, Bobik E, Leonard AE, Parker-Barnes JM, Huang YS, Mukerji P (2000) Polyunsaturated fatty acid-specific elongation enzymes. Biochem Soc Trans 28:658–660
Domergue F, Lerchl J, Zähringer U, Heinz E (2002) Cloning and functional characterization of Phaeodactylum tricornutum front-end desaturases involved in eicosapentaenoic acid biosynthesis. Eur J Biochem 269:4105–4113
Donot F, Fontana A, Baccou JC, Strub C, Schorr-Galindo S (2014) Single cell oils (SCOs) from oleaginous yeasts and moulds: production and genetics. Biomass Bioenergy 68:135–150
Fillet S, Ronchel C, Callejo C, Fajardo MJ, Moralejo H, Adrio JL (2017) Engineering Rhodosporidium toruloides for the production of very long-chain monounsaturated fatty acid-rich oils. Appl Microbiol Biotechnol 101:7271–7280
Fujiwara D, Yoshimoto H, Sone H, Harashima S, Tamai Y (1998) Transcriptional co-regulation of Saccharomyces cerevisiae alcohol acetyltransferase gene, ATF1 and Δ-9 fatty acid desaturase gene, OLE1 by unsaturated fatty acids. Yeast 14:711–721
Jiang M, Guo B, Wan X, Gong Y, Zhang Y, Hu C (2014) Isolation and characterization of the diatom Phaeodactylum Δ5-elongase gene for transgenic LC-PUFA production in Pichia pastoris. Mar Drugs 12:1317–1334
Juanssilfero AB, Kahar P, Amza RL, Miyamoto N, Otsuka H, Matsumoto H, Kihira C, Thontowi A, Ogino C, Prasetya B, Kondo A (2018) Effect of inoculum size on single-cell oil production from glucose and xylose using oleaginous yeast Lipomyces starkeyi. J Biosci Bioeng 125:695–702
Kainou K, Kamisaka Y, Kimura K, Uemura H (2006) Isolation of Δ12 and ω3-fatty acid desaturase genes from the yeast Kluyveromyces lactis and their heterologous expression to produce linoleic and α-linolenic acids in Saccharomyces cerevisiae. Yeast 23:605–612
Kamisaka Y, Kimura K, Uemura H, Shibakami M (2010) Activation of diacylglycerol acyltransferase expressed in Saccharomyces cerevisiae: overexpression of Dga1p lacking the N-terminal region in the Δsnf2 disruptant produces a significant increase in its enzyme activity. Appl Microbiol Biotechnol 88:105–115
Kamisaka Y, Kimura K, Uemura H, Yamaoka M (2013) Overexpression of the active diacylglycerol acyltransferase variant transforms Saccharomyces cerevisiae into an oleaginous yeast. Appl Microbiol Biotechnol 97:7345–7355
Kobayashi SD, Nagiec MM (2003) Ceramide/long-chain base phosphate rheostat in Saccharomyces cerevisiae: regulation of ceramide synthesis by Elo3p and Cka2p. Eukaryot Cell 2:284–294
Kosa M, Ragauskas AJ (2011) Lipids from heterotrophic microbes: advances in metabolism research. Trends Biotechnol 29:53–61
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25:402–408
Matsuzawa T, Maehara T, Kamisaka Y, Ara S, Takaku H, Yaoi K (2018) Identification and characterization of Δ12 and Δ12/Δ15 bifunctional fatty acid desaturases in the oleaginous yeast Lipomyces starkeyi. Appl Microbiol Biotechnol 102:8817–8826
Möller S, Croning MD, Apweiler R (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646–653
Nakagawa Y, Sakumoto N, Kaneko Y, Harashima S (2002) Mga2p is a putative sensor for low temperature and oxygen to induce OLE1 transcription in Saccharomyces cerevisiae. Biochem Biophys Res Commun 291:707–713
Nakagawa Y, Ueda A, Kaneko Y, Harashima S (2003) Merging of multiple signals regulating Δ9 fatty acid desaturase gene transcription in Saccharomyces cerevisiae. Mol Gen Genomics 269:370–380
Oguro Y, Yamazaki H, Ara S, Shida Y, Ogasawara W, Takagi M, Takaku H (2017) Efficient gene targeting in non-homologous end-joining-deficient Lipomyces starkeyi strains. Curr Genet 63:751–763
Oh CS, Toke DA, Mandala S, Martin CE (1997) ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem 272:17376–17384
Parker-Barnes JM, Das T, Bobik E, Leonard AE, Thurmond JM, Chaung LT, Huang YS, Mukerji P (2000) Identification and characterization of an enzyme involved in the elongation of n-6 and n-3 polyunsaturated fatty acids. Proc Natl Acad Sci U S A 97:8284–8289
Prasitchoke P, Kaneko Y, Sugiyama M, Bamba T, Fukusaki E, Kobayashi A, Harashima S (2007) Functional analysis of very long-chain fatty acid elongase gene, HpELO2, in the methylotrophic yeast Hansenula polymorpha. Appl Microbiol Biotechnol 76:417–427
Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, Göker M, Salamov AA, Wisecaver JH, Long TM, Calvey CH, Aerts AL, Barry KW, Choi C, Clum A, Coughlan AY, Deshpande S, Douglass AP, Hanson SJ, Klenk HP, LaButti KM, Lapidus A, Lindquist EA, Lipzen AM, Meier-Kolthoff JP, Ohm RA, Otillar RP, Pangilinan JL, Peng Y, Rokas A, Rosa CA, Scheuner C, Sibirny AA, Slot JC, Stielow JB, Sun H, Kurtzman CP, Blackwell M, Grigoriev IV, Jeffries TW (2016) Comparative genomics of biotechologically important yeasts. Proc Natl Acad Sci U S A 113:9882–9887
Rössler H, Rieck C, Delong T, Hoja U, Schweizer E (2003) Functional differentiation and selective inactivation of multiple Saccharomyces cerevisiae genes involved in very-long-chain fatty acid synthesis. Mol Gen Genomics 269:290–298
Schneiter R, Tatzer V, Gogg G, Leitner E, Kohlwein SD (2000) Elo1p-dependent carboxy-terminal elongation of C14:1Δ9 to C16:1Δ11 fatty acids in Saccharomyces cerevisiae. J Bacteriol 182:3655–3660
Stukey JE, McDonough VM, Martin CE (1990) The OLE1 gene of Saccharomyces cerevisiae encodes the Δ9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem 265:20144–20149
Sutanto S, Zullaikah S, Tran-Nguyen PL, Ismadji S, Ju YH (2018) Lipomyces starkeyi : its current status as a potential oil producer. Fuel Process Technol 177:39–55
Toke DA, Martin CE (1996) Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J Biol Chem 271:18413–18422
Yamazaki H, Kobayashi S, Ebina S, Abe S, Ara S, Shida Y, Ogasawara W, Yaoi K, Araki H, Takaku H (2019) Highly selective isolation and characterization of Lipomyces starkeyi mutants with increased production of triacylglycerol. Appl Microbiol Biotechnol 103:6297–6308
Zimmermann C, Santos A, Gable K, Epstein S, Gururaj C, Chymkowitch P, Pultz D, Rødkær SV, Clay L, Bjørås M, Barral Y, Chang A, Færgeman NJ, Dunn TM, Riezman H, Enserink JM (2013) TORC1 inhibits GSK3-mediated Elo2 phosphorylation to regulate very long chain fatty acid synthesis and autophagy. Cell Rep 27:1036–1046
Funding
This study was supported by the New Energy and Industrial Technology Development Organization (NEDO).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Matsuzawa, T., Kamisaka, Y., Maehara, T. et al. Identification and characterization of two fatty acid elongases in Lipomyces starkeyi. Appl Microbiol Biotechnol 104, 2537–2544 (2020). https://doi.org/10.1007/s00253-020-10401-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10401-9