Abstract
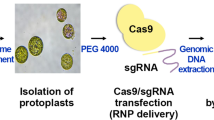
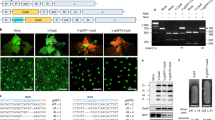
The CRISPR–Cas9 system is a powerful tool for editing genes of interest in specific plant genomes. Indeed, genome-editing systems have been used to enhance a variety of agricultural traits and to study gene functions in many plant species, and the plasmid-mediated delivery of Cas9 and single guide RNA (sgRNA) to plants has been reported to facilitate highly efficient editing. However, the random and stable integration of plasmid DNA sequences into plant genomes can cause insertional mutagenesis, and an additional step is required to remove such foreign sequences from edited plant genomes. Accordingly, the aim of the present study was to investigate the effectiveness of directly delivering purified CRISPR–Cas9 ribonucleoproteins (RNPs) to protoplasts from cabbage (Brassica oleracea var. capitata), an important cruciferous vegetable. The flowering-time regulator gene GIGANTEA (GI) was targeted, with the goal of delaying flowering time and prolonging vegetative growth. We investigated the targeted mutagenesis insertion and deletion rates using targeted deep sequencing. The mutation frequency achieved using one of the sgRNAs (sgRNA2) was 2% in the infected protoplast. The shoots were regenerated from 44% (46/103) of protoplast-derived calli. Consequently, three independent and completely transgene-free mutants were obtained, including one homogeneous biallelic line in which both GI alleles were successfully edited, thereby yielding a complete GI knockout line. These results suggest that the transgene-free CRISPR–Cas9 system is a promising tool for improving agricultural beneficial traits of cabbage.




Similar content being viewed by others
References
Andersson M, Turesson H, Olsson N, Fält A-S, Ohlsson P, Gonzalez MN, Samuelsson M, Hofvander P (2018) Genome editing in potato via CRISPR–Cas9 ribonucleoprotein delivery. Physiol Plant 164(4):378–384
Bradford KJ, Van Deynze A, Gutterson N, Parrott W, Strauss SH (2005) Regulating transgenic crops sensibly: lessons from plant breeding, biotechnology and genomics. Nat Biotechnol 23(4):439–444
Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70(1):667–697
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186(2):757–761
Gaj T, Gersbach CA, Barbas CF (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31(7):397–405
Gerszberg A (2018) Tissue culture and genetic transformation of cabbage (Brassica oleracea var. capitata): an overview. Planta 248(5):1037–1048
Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA (1983) A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303(5913):179–180
Hsu PD, Lander Eric S, Zhang F (2014) Development and applications of CRISPR–Cas9 for genome engineering. Cell 157(6):1262–1278
Jie EY, Kim SW, Jang HR, In DS, Liu JR (2011) Myo-inositol increases the plating efficiency of protoplast derived from cotyledon of cabbage (Brassica oleracea var. capitata). J Plant Biotechnol 38(1):69
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821
Kim SY, Park BS, Kwon SJ, Kim J, Lim MH, Park YD, Kim DY, Suh SC, Jin YM, Ahn JH, Lee YH (2007) Delayed flowering time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Cell Rep 26(3):327–336
Kim WY, Ali Z, Park HJ, Park SJ, Cha JY, Perez-Hormaeche J, Quintero FJ, Shin G, Kim MR, Qiang Z, Ning L, Park HC, Lee SY, Bressan RA, Pardo JM, Bohnert HJ, Yun DJ (2013) Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat Commun 4:1352
Lawrenson T, Shorinola O, Stacey N, Li C, Østergaard L, Patron N, Uauy C, Harwood W (2015) Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol 16(1):258
Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, Gao C (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8:14261
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IAP, Zhao M, Ma J, Yu J, Huang S, Wang X, Wang J, Lu K, Fang Z, Bancroft I, Yang T-J, Hu Q, Wang X, Yue Z, Li H, Yang L, Wu J, Zhou Q, Wang W, King GJ, Pires JC, Lu C, Wu Z, Sampath P, Wang Z, Guo H, Pan S, Yang L, Min J, Zhang D, Jin D, Li W, Belcram H, Tu J, Guan M, Qi C, Du D, Li J, Jiang L, Batley J, Sharpe AG, Park B-S, Ruperao P, Cheng F, Waminal NE, Huang Y, Dong C, Wang L, Li J, Hu Z, Zhuang M, Huang Y, Huang J, Shi J, Mei D, Liu J, Lee T-H, Wang J, Jin H, Li Z, Li X, Zhang J, Xiao L, Zhou Y, Liu Z, Liu X, Qin R, Tang X, Liu W, Wang Y, Zhang Y, Lee J, Kim HH, Denoeud F, Xu X, Liang X, Hua W, Wang X, Wang J, Chalhoub B, Paterson AH (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5:3930
Ma C, Liu M, Li Q, Si J, Ren X, Song H (2019a) Efficient BoPDS gene editing in cabbage by the CRISPR/Cas9 system. Hortic Plant J 5(4):164–169
Ma C, Zhu C, Zheng M, Liu M, Zhang D, Liu B, Li Q, Si J, Ren X, Song H (2019b) CRISPR/Cas9-mediated multiple gene editing in Brassica oleracea var. capitata using the endogenous tRNA-processing system. Hortic Res 6(1):20
Malnoy M, Viola R, Jung MH, Koo OJ, Kim S, Kim JS, Velasco R, Nagamangala Kanchiswamy C (2016) DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci 7:1904
Mishra P, Panigrahi KC (2015) GIGANTEA—an emerging story. Front Plant Sci 6:8
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497
Murovec J, Guček K, Bohanec B, Avbelj M, Jerala R (2018) DNA-free genome editing of Brassica oleracea and B. rapa protoplasts using CRISPR–Cas9 ribonucleoprotein complexes. Front Plant Sci 9:1594
Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A (2016) Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nature Commun 7:13274
Woo JW, Kim J, Kwon SI, Corvalán C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS (2015) DNA-free genome editing in plants with preassembled CRISPR–Cas9 ribonucleoproteins. Nat Biotechnol 33:1162
Wright DA, Townsend JA, Winfrey RJ Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF (2005) High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J 44(4):693–705
Xie Q, Lou P, Hermand V, Aman R, Park HJ, Yun D-J, Kim WY, Salmela MJ, Ewers BE, Weinig C, Khan SL, Schaible DLP, McClung CR (2015) Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc Natl Acad Sci USA 112(12):3829–3834
Xiong X, Liu W, Jiang J, Xu L, Huang L, Cao J (2019) Efficient genome editing of Brassica campestris based on the CRISPR/Cas9 system. Mol Genet Genom. https://doi.org/10.1007/s00438-019-01564-w
Yang H, Wu JJ, Tang T, Liu KD, Dai C (2017) CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci Rep 7(1):7489
Acknowledgements
This research was supported by grants from the KRIBB Initiative Program, Next-Generation BioGreen 21 Program (SSAC) and Rural Development Administration, Republic of Korea (Grant No. PJ01318604).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Park, SC., Park, S., Jeong, Y.J. et al. DNA-free mutagenesis of GIGANTEA in Brassica oleracea var. capitata using CRISPR/Cas9 ribonucleoprotein complexes. Plant Biotechnol Rep 13, 483–489 (2019). https://doi.org/10.1007/s11816-019-00585-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-019-00585-6