Abstract
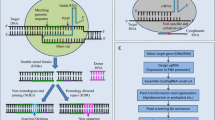
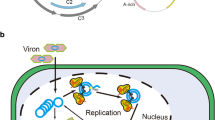
The CRISPR–Cas9 is emerging genome editing tool and very easy and straightforward in operation that has been tested and explored for introduction of new traits in plant systems. Recently, a number of reports have documented utilization of this technology for providing tolerance against viral diseases mediated by begomoviruses. Begomoviruses infect dicot and are transmitted by white flies and cause devastating losses to yield of important agricultural crops including tomato, cassava and cotton. An overview of genomic structure of begomoviruses has been presented to understand the potential strategy for designing of effective sgRNAs to combat the viral replication for generating resistance against infection. This review provides the introduction, recent developments, and applications of the CRISPR–Cas9 system in plants and proposes a holistic methodology for generating cotton plant an example having resistance against begomoviruses. The genome editing using CRISPR–Cas9 system against complex of begomoviruses collectively termed as cotton leaf curl virus, which a major contributor to reduction of the cotton yield especially in Northern India and Pakistan is also discussed thoroughly. In conclusion, this potential strategy could be a sustainable approach for development of tolerant crops against diseases mediated by DNA viruses.




Similar content being viewed by others
Abbreviations
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- Cas:
-
CRISPR associated protein
- sgRNA:
-
Single guide RNA
- crRNA:
-
CRISPR RNA
- PAM:
-
Protospacer adjacent motif
- CLCuV:
-
Cotton leaf curl virus
- tracRNA:
-
Trans activating RNA
- NW:
-
New world
- OW:
-
Old world
- CLCuD:
-
Cotton leaf curl disease
References
Ahmad S, Mahmood K, Hanif M, Nazeer W, Malik W, Qayyum A et al (2011) Introgression of cotton leaf curl virus-resistant genes from Asiatic cotton (Gossypium arboretum) into upland cotton (G. hirsutum). Genet Mol Res 10:2404–2414
Ahuja SL, Monga D, Dhayal LS (2007) Genetics of resistance to cotton leaf curl disease in Gossypiumhirsutum L. under field conditions. J Hered 98:79–83
Ali I, Amin I, Briddon RW, Mansoor S (2013) Artificial microRNA-mediated resistance against the monopartite begomovirus Cotton leaf curl Burewala virus. Virol J 10:231
Ali Z, Abul-faraj A, Idris A, Ali S, Tashkandi M, Mahfouz M (2015a) CRISPR/Cas9-mediated viral interference in plants. Genome Biol 16:238
Ali Z, Abul-Faraj A, Li L, Ghosh N, Piatek M, Mahjoub A et al (2015b) Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant. https://doi.org/10.1016/j.molp.2015.02.011
Amudha J, Balasubrami G, Malathi VG, Monga D, Kranthi KR (2011) Cotton leaf curl virus resistance transgenic with antisense coat protein gene (AV1). Curr Sci 101:300–307
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26:151–163
Bassett AR, Tibbit C, Ponting CP, Liu JL (2013) Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 4:220–228
Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013) Plant genome editng made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:39
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52
Caroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188:773–782
Chen W, Qian Y, Wu X, Sun Y, Wu X, Cheng X (2014) Inhibiting replication of begomoviruses using artificial zinc finger nucleases that target viral-conserved nucleotide motif. Virus Genes 48:494–501
Cheng X, Li F, Cai J, Chen W, Zhao N, Sun Y et al (2015) Artificial TALE as a convenient protein platform for engineering board spectrum resistance to begoviruses. Viruses 7:4772–4782
Cho SW, Kim S, Kim JM, Kim JS (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. NatBiotechnol 31:230–232
Cong L, Ran FA, Cox D, Lin S, Barreto R, Habib N et al (2013) Multiplex genome engineering using CRISPR/Cas system. Science 339:819–823
Farooq A, Farooq J, Mahmood A, Shakeel A, Rehman A, Batool A et al (2011) An overview of cotton leaf curl virus disease (CLCuD) a serious threat to cotton productivity. Aust J Crop Sci 13:182301831
Fauser F, Roth N, Pacher M, Ilg G, Sanchez-Fernandez R, Biesgen C et al (2012) In planta gene targeting. Proc Natl Acad Sci USA 109:7535–7540
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P et al (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23:1229–1232
Feng Y, Zhang S, Huang X (2014) A robust TALENS system for highly efficient mammalian genome editing. Sci Rep 4:3632
Gao W, Long L, Tian X et al (2017) Genome editing in cotton with the CRISPR/Cas9 system. Front Plant Sci 8:1364
Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109:E2579–E2586
Hanley-Bowdoin L, Settlage S, Orozco BM, Nagar S, Robertson D (1999) Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit Rev Plant Sci 18:71–106
Hanley-Bowdoin L, Bejarano ER, RobertsonD MS (2013) Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11:777–788
Heigwer F, Zhan T, Breinig M, Winter J, Brṻgermann D, Leible S, Boutros M (2016) CRISPR library designer (CLD): software for multiplexing design of single guide RNA libraries. Genome Biol 24:55
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agalwala V (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. NatBiotechnol 31:827–832
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD et al (2013) Efficient genome editing in zebrafish using a CRISPR–Cas system. Nat Biotechnol 31:227–229
Ji X, Zhang H, Zhang Y, Wang Y, Gao C (2015) Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat Plants 1:15144
Jia H, Wang N (2014) Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9:e93806
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2014) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e188
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Johnson RA, Gurevich V, Filler S, Samach A, Levy A (2015) Comparative assessments of CRISPR-Casnucleases’s cleavage efficiency in planta. Plant Mol Biol 87:143–156
Koshino-Kimura Y, Takenaka K, Domoto F, Ohashi M, Miyazaki T, Aoyama Y et al (2009) Construction of plants resistant to TYLCV by using artificial zinc-finger proteins. Nucleic Acids Symp Ser (Oxf) 53:281–282
Liang Z, Zhang K, Chen K, Gao C (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genom 41:63–68
Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE et al (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Mansoor S, Briddon RW, Zafar Y, Stanley J (2003) Geminivirus disease complexes: an emerging threat. Trends Plant Sci 8:128–134
Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol Plant 6:2008–2011
Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X et al (2013) Targeted mutagenesis in rice using CRISPR–Cas system. Cell Res 23:1233–1236
Mikami M, Toki S, Endo M (2015) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88:561–572
Mori T, Takenaka K, Domoto F, Aoyama Y, Sera T (2013) Inhibition of binding of tomato yellow leaf curl virus rep to its replication origin by artificial zinc-finger protein. Mol Biotechnol 54:198–203
Nawaz-ul-Rehman MS, Fauquet CM (2009) Evolution of geminiviruses and their satellites. FEBS Lett 583:1825–1832
Nazeer W, Tipu AL, Ahmad S, Mahmood K, Mahmood A, Zhou B (2014) Evaluation of cotton leaf curl virus resistance in BC1, BC2, and BC3 progenies from an interspecific cross between Gossypiumarboreum and Gossypiumhirsutum. PLOS One. https://doi.org/10.1371/journal.pone.0111861
Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L et al (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156:836–843
OECD/FAO (2018) OECD-FAO agricultural outlook 2018–2027. OECD Publishing, Food and Agriculture Organization of the United Nations, Paris, Rome
Park J, Kin JS, Bae S (2016) Cas-Database: web-based genome-wide guide RNA library design for gene knockout screens using CRISPR–Cas9. Bioinformatics. https://doi.org/10.1093/bioinformatics/btw103
Qazi J, Amin I, Mansoor S, Iqbal MJ, Briddon RW (2007) Contribution of the satellite encoded gene βC1 to cotton leaf curl disease symptoms. Virus Res 128:135–139
Schiffer JT, Aubert M, Weber ND, Mintzer E, Stone D, Jerome KR (2012) Targeted DNA mutagenesis for the cure of chronic viral infections. J Virol 86:8920–8936
Sera T (2015) Inhibition of virus DNA replication by artificial zinc finger proteins. J Virol 79:2614–2619
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z et al (2013) Targeted genome modification of crop plants using a CRISPR–Cas system. Nat Biotechnol 31:686–688
Sohrab SS, Kamal MA, Llah A, Husen A, Bhattacharya PS, Rana D (2014) Development of cotton leaf curl virus resistant transgenic cotton using antisense βC1. Saudi J Biol Sci 23:358–362
Suenaga T, Kohyama M, Hirayasu K, Arase H (2014) Engineering large viral DNA genomes using the CRISPR–Cas9 system. Microbiol Immunol 58:513–522
Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I et al (2014) CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55:475–481
Upadhyay SK, Kumar J, Alok A, Tuli R (2013) RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 3:2233–2238
Varma A, Malathi V (2003) Emerging gemivirus problems: a series threat to crop production. Ann Appl Biol 142:145–164
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C et al (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32:947–951
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR–Cas system. Mol Plant 6:1975–1983
Yang H, Wang H, Jaenisch R (2014) Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 9:1956–1968
Yin K, Han T, Liu G, Chen T, Wang Y, Yu AYL et al (2015) Ageminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genme editing. Sci Rep 5:14926
Yuen KS, Chan CP, Wong NH, Ho CH, Ho TH, Lei T et al (2015) CRISPR/Cas9-mediated genome editing of Epstein–Barr virus in human cells. J Gen Virol 96:626–636
Zebrini FM, Briddon RW, Irdis A et al (2017) ICTV virus taxonomy profile: geminiviridae. J Gen Virol 98:131–133
Zhen S, Hua L, Liu Y-H, Gao L-C, Fu J et al (2015) Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther 22:404–412
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42:10903–10914
Acknowledgements
Authors thank the anonymous reviewers for their critical comments and suggestions. The authors also would like to express deepest gratitude to Vice Chancellor of Central University of Punjab, India for providing unconditional supports during course of this work. This research is sponsored by “Research Seed Money” grant as sanctioned by Central University of Punjab, India.
Author information
Authors and Affiliations
Contributions
VK and SKY conceived and designed present work. VK and SKY analyzed data. APU and VK wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Uniyal, A.P., Yadav, S.K. & Kumar, V. The CRISPR–Cas9, genome editing approach: a promising tool for drafting defense strategy against begomoviruses including cotton leaf curl viruses. J. Plant Biochem. Biotechnol. 28, 121–132 (2019). https://doi.org/10.1007/s13562-019-00491-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-019-00491-6