Abstract
Quantifying the inner structure of bones is central to various analyses dealing with the phenotypic evolution of animals with an ossified skeleton. Computed tomography allows to assess the repartition of bone tissue within an entire skeletal element. Two parameters of importance for such analyses are the global compactness (Cg) and total cross-sectional area (Tt.Ar). However, no open-source, time-efficient methods are available to acquire these parameters for whole bones. A methodology to assess the variation of these parameters along a profile following one of the studied bone’s anatomical axes is also wanting. Here I present an ImageJ macro and associated R script to automatically acquire Cg and Tt.Ar along an axis of the skeletal element of interest using a slice-by-slice approach. No manual segmentation is required and several bones can be present on the analysed scan, as long as the bone of interest is isolated and the largest element on each slice. While some bias might be involved by the automatic acquisition, semi-automatic slice exclusion and correction procedures can be used to efficiently account for it. As a test case, µCT-data was gathered for the mid-lumbar vertebra of over 70 mammals. The two evaluated correction procedures proved to perform equally well, with a slight advantage for the one relying on the exclusion of local outliers. The presented macro allows to efficiently build a dataset concerned with the quantification of bone inner structure. The code being readily available, further improvement of the methodology and adjustment to particular needs can be easily performed.
Similar content being viewed by others
Introduction
Assessing the inner structure of bones is a critical aspect of many analyses dealing with the phenotype of animals with an ossified skeleton. Indeed, quantifying the repartition of bone tissue within skeletal elements has been for instance employed for functional anatomy (e.g., Houssaye and Botton-Divet 2018) (palaeo) biology and (palaeo) ecology (e.g., Laurin et al. 2004), or taxonomy (Amson et al. 2015) studies. While the first analyses quantifying bone structure used physical sections (e.g., Quekett 1849), which were hence limited to a particular region of the studied bone, the popularization of Computed tomography (CT) allowed for the anatomy of whole skeletal elements to be studied. Given that CT-scans are routinely reconstructed as a stack of two-dimensional (2D) images, several methods involve a slice-by-slice approach to quantify bone structure along an anatomical axis of the studied skeletal element. Such an approach is for instance implemented in the plugin BoneJ (Doube et al. 2010), for ImageJ (Schneider et al. 2012). The ‘Slice geometry’ routine of this plugin performs measurements (e.g., cross-sectional area, second moment of area around major and minor axes, maximum thickness in 2D) on all foreground pixels of the studied slices. The commonly acquired parameter referred to as global compactness (Cg), or ratio of cross-sectional area occupied by bone divided by whole cross-sectional area is not acquired by the latter routine. This parameter, along with the 3D equivalent referred to as bone fraction (BV.TV), have been shown to preponderantly explain the mechanical properties of bones (Musy et al. 2017) and are clearly associated with evolutionary adaptations (de Buffrénil et al. 2010). These parameters are hence routinely acquired in various studies dealing with bone structure function (e.g., Ryan and Ketcham 2002) and more generally lifestyle (e.g., Laurin et al. 2004). The acquisition of these parameters requires the definition of a region of interest. In 2D, this often corresponds to the total cross-sectional area (Tt.Ar), i.e., a selection encompassing the skeletal element and the internal vacuities it comprises. Such a selection is not trivial in some cases, in particular when porosity connects the area corresponding to the exterior of the bone to large internal vacuity (see Methods S1 in Supplementary Material 3). For this purpose, previous studies have resorted to manually or automatically segmenting the skeletal element with other (commercial, closed-source) softwares (Behrooz et al. 2017; Gross et al. 2014; Houssaye et al. 2018). It is noteworthy that the free but closed-source software Bone Profiler (Girondot and Laurin 2003), implements an automatic global compactness acquisition. But the latter is restricted to acquiring parameters for single sections, and large porosity linking the outside of the bone to its internal vacuities, if present, will bias the output.
The methodology presented herein was designed to provide an open-source, time-efficient solution to acquire Cg and Tt.Ar of whole skeletal elements for large (comparative) datasets. No segmentation of the element of interest is required. Several elements can be present in the studied stack, as long as the Tt.Ar value of the element of interest is the highest on each slice. Data can therefore be directly acquired from raw scan stacks.
Functionality
ImageJ macro
The macro (see Data Accessibility section and Methods S1 in Supplementary Material 3) is written in the ImageJ macro language, and is therefore intended to be run by ImageJ (Schneider et al. 2012) or Fiji (Schindelin et al. 2012). The macro should be applied to a stack comprising the element of interest, oriented consistently. For example, for vertebrae, the anteroposterior axis can be aligned to the Z-axis of the stack (i.e., direction of slice superimposition). For long bones, it is the proximodistal axis that will typically be aligned to the Z-axis, in order to analyse transverse cross-sections. The stack can either be in greyscale (thresholding would be performed with the ‘Optimise Threshold > Threshold Only’ routine of BoneJ; Doube et al. 2010) or already binarized (foreground: bone pixels; background, non-bone pixels). The unit of pixel length (often already a property of the image stack) is expected to be either mm or inch (the latter will be converted into mm).
The macro proceeds slice by slice. First it selects an area corresponding to the bone of interest, assuming that it is the largest object of the slice, with the ‘Analysing Particles’ routine of ImageJ. Small undesired particles possibly present in the stack are excluded from to process to speed it up (minimum analysed particle area set by default to square of 10% of width of whole slice). Tt.Ar (area of selection) and Cg (percent of selection area occupied by bone) are then measured (‘Measure’ routine of ImageJ). Finally, the macro allows the user to specify which are the first and last slices of interest, and also whether there are slices to exclude from the analysis (see below). The output is a result table where the rows correspond to slices, the variable ‘ResArea’ corresponds to Tt.Ar, the variable ‘ResC’ corresponds to the global compactness, and the variable ‘ToDel’ corresponds to the status of the slice: ‘Z1’ and ‘Z2’, first and last slice of interest, respectively; ‘0’, slice to include; ‘1’, slice to exclude. The mean Tt.Ar and mean Cg (of all slices from Z1 to Z2 but the excluded ones) are also provided in the Log window, along with a result plot window showing the global compactness profile.
For some slices, porosity can connect the area corresponding to the exterior of the bone to large internal vacuity (e.g., the vertebral canal of a vertebra, see Methods S1 in Supplementary Material 3). In that case, the automated selection is flawed, and the corresponding measurements should be excluded for subsequent analysis. Depending on the number of slices to exclude and the scanning resolution, experience has shown that identifying all of these slices can be difficult. Therefore, it is recommended to perform additional procedures in order to account for the potential bias associated with overlooked slices. Two examples of such procedures are included in the accompanying R script (see below).
Profile Correction Procedures
The accompanying script (see Data Accessibility section), written with R v. 3.5.1 (R Core Team 2018), provides an example of procedure to load data as produced by the ImageJ macro described above. In addition to loading a series of data files (each corresponding to a specimen), this script replaces the parameter values of the excluded slices by a mean of those of their neighbours (or a sequence of values when several subsequent slices are concerned).
The script also includes two procedures dealing with measurements flawed by incorrect automatic selection (see above). The first procedure defines local outliers, which typically correspond to a single slice or a short sequence of slices for which automatic selection was not performed accurately. These local outliers are defined as slices for which the measured compactness value exceeds the 75th percentile plus 1.5 times the interquartile range of a subsample of the six neighbouring slices’ values excluding the direct neighbours of the slice of interest (which have greater chances of also being affected by an incorrect automatic selection). The second procedure is designed to reduce the impact of those slices for which automatic selection was not performed accurately using curve smoothing approach. It involves performing a loess regression on all slices except those that were manually excluded (during the run of the ImageJ macro). This non-parametric local polynomial regression uses a nearest-neighbour approach. The loess function (base of R) was used with a span of 1 (all the stack is included in the neighbourhood used for the local fit), in order to strongly smooth the profile curve.
Vertebra Scans
As a test sample, a dataset consisting of 71 therian mammals of various body sizes and of lifestyles was built. Specimens were selected to be adult, wild-caught, and free of apparent diseases. The vertebra from the middle of the lumbar series was sampled. The vertebrae were µCT-scanned whole, either isolated or articulated to the rest of the skeleton. Depending on the size and geometry of the vertebrae, resolution ranged from 3.3 to 98 µm. Scans were acquired with a Phoenix nanotom (General Electric GmbH Wunstorf, Germany) and a FF35-CT-System (YXLON GmbH Hamburg, Germany). In the case of vertebrae, the first and last slice of interest corresponds respectively to the anterior-most and posterior-most slices for which the vertebral canal is complete (see above and Methods S1 in Supplementary Material 3).
Limitations
While no segmentation is needed, the methodology requires for the bone of interest to be the largest element on each slice. Performing a quick cropping of the bone of interest –this can be done in ImageJ making a rectangular selection and using the ‘Duplicate’ routine selecting the appropriate range of slices—is usually enough to obtain such a disposition. If large unwanted elements remain, they can be quickly deleted from the slices of interest by filling an appropriate selection with the background colour (this can be efficiently done on multiple slices using the ‘Interpolate ROIs’ and ‘Fill’ routines of the ROI manager). Furthermore, if after thresholding other bones are connected to the bone of interest, there will be no other solution than to resort to manual segmentation.
Example Application: Lumbar Vertebrae in Therian Mammals
Process time of the macro was 78 s for the largest stack (12.6 GB; with a 2.53 GHz processor). Two different data acquisitions were performed. One for which the stacks were carefully inspected, in order to manually exclude as many slices for which automatic selection was biased as possible. For the second ‘rough’ acquisition, only a quick manual exclusion of the latter slices was performed. Comparison of two procedures for correcting profiles yielded very similar results (Table 1). The one using local outliers was found as slightly more accurate than the one based on loess regression. The former is hence recommended (note that the differences between the correction procedures and not applying any procedure were very minor; Table 1), and only the parameters resulting from this procedure will be further presented.
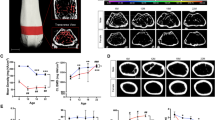
For the mean Cg, values range from 15% (Pipistrellus pipistrellus, common pipistrelle; Fig. 1a) to 72% (Dugon dugon, dugong; Fig. 1b), with an average mean value of 34% (approached by Echimys chrysurus, white-faced spiny tree-rat; Fig. 1c), and an average standard deviation of 3%. A diverse range of values is described by the other therian mammals sampled (Fig. 2).
Examples of global compactness profiles (Cg). a Least compact vertebra (Pipistrellus pipistrellus, ZMB Mam 55128). b Most compact vertebra (Dugon dugon, ZMB Mam 69340). c Vertebra with a value close to the average of the dataset (Echimys chrysurus, ZMB Mam 8345). ‘Z1’ and ‘Z2’ define the first and last slices of interest, respectively. Orange circles denote excluded slices (mean of neighbours’ values were used as replacement). Loess regression curve is in red
Slice-by-slice quantification allows the user to assess the evolution of the parameters along the profile of the studied element. The shape of the Cg profile is quite uniform across the dataset. For most of the specimens sampled, Cg values are relatively high anteriorly, slightly increase in the anterior region of the profile, then decrease in the central region of the vertebra, and finally increase again in the posterior region, forming a sigmoid curve (Fig. 1b, c). The central region of the centrum is marked by relatively large vascular canals for most of the specimens (see excluded series of slices around position ‘330’ and ‘605’ for Fig. 1b and c, respectively). It is therefore recommended to inspect with particular care this region in order to identify the slices to be excluded from the analysis.
Conclusion
The ImageJ macro and associated R script presented herein offer an open-source, time-efficient methodology to quantify the global compactness and total cross-sectional area along a bone’s anatomical axis. As a test case, a dataset comprising mid-lumbar vertebra scans for over 70 mammals was acquired. It is shown that the automated method involves some biased measurements. But the latter can be minimized using a semi-automated exclusion of slices and a correction protocol. The corrected profiles, representing in this case the variation of global compactness or total cross-sectional area along the anteroposterior axis of the vertebra, can readily be compared across the diversity of the clade of study.
Data Accessibility
Both the ImageJ macro and R script are available on GitHub, https://github.com/eliamson/sliceByslice/.
References
Amson, E., de Muizon, C., Domning, D. P., Argot, C., & de Buffrénil, V. (2015). Bone histology as a clue for resolving the puzzle of a dugong rib in the Pisco Formation, Peru. Journal of Vertebrate Paleontology, 35(3), e922981.
Behrooz, A., Kask, P., Meganck, J., & Kempner, J. (2017). Automated quantitative bone analysis in in vivo x-ray micro-computed tomography. IEEE Transactions on Medical Imaging, 36(9), 1955–1965.
de Buffrénil, V., Canoville, A., D’Anastasio, R., & Domning, D. P. (2010). Evolution of sirenian pachyosteosclerosis, a model-case for the study of bone structure in aquatic tetrapods. Journal of Mammalian Evolution, 17(2), 101–120.
Doube, M., Kłosowski, M. M., Arganda-Carreras, I., Cordelières, F. P., Dougherty, R. P., Jackson, J. S., et al. (2010). BoneJ: Free and extensible bone image analysis in ImageJ. Bone, 47(6), 1076–1079.
Girondot, M., & Laurin, M. (2003). Bone profiler: a tool to quantify, model, and statistically compare bone-section compactness profiles. Journal of Vertebrate Paleontology, 23(2), 458–461.
Gross, T., Kivell, T. L., Skinner, M. M., Nguyen, N. H., & Pahr, D. H. (2014). A CT-image-based framework for the holistic analysis of cortical and trabecular bone morphology. Palaeontologia Electronica, 17(3), 1–13.
Houssaye, A., & Botton-Divet, L. (2018). From land to water: Evolutionary changes in long bone microanatomy of otters (Mammalia: Mustelidae). Biological Journal of the Linnean Society, 125(2), 240.
Houssaye, A., Taverne, M., & Cornette, R. (2018). 3D quantitative comparative analysis of long bone diaphysis variations in microanatomy and cross-sectional geometry. Journal of Anatomy, 25(5), 836–849.
Kumar, S., Stecher, G., Suleski, M., & Hedges, S. B. (2017). TimeTree: A resource for timelines, timetrees, and divergence times. Molecular biology and evolution, 34(7), 1812–1819.
Laurin, M., Girondot, M., & Loth, M.-M. (2004). The evolution of long bone microstructure and lifestyle in lissamphibians. Paleobiology, 30(4), 589–613.
Musy, S. N., Maquer, G., Panyasantisuk, J., Wandel, J., & Zysset, P. K. (2017). Not only stiffness, but also yield strength of the trabecular structure determined by non-linear µFE is best predicted by bone volume fraction and fabric tensor. Journal of the Mechanical Behavior of Biomedical Materials, 65, 808–813.
Quekett, J. (1849). On the intimate structure of bone, as composing the skeleton, in the four great classes of animals, viz, mammals, birds, reptiles, and fishes, with some remarks on the great value of the knowledge of such structure in determining the affinities of minute. Transactions of the Microscopical Society of London, 2(1), 46–58.
R Core Team. (2018). R: A language and environment for statistical computing. Vienna: R foundation for statistical computing.
Revell, L. J. (2012). phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3(2), 217–223.
Ryan, T. M., & Ketcham, R. A. (2002). The three-dimensional structure of trabecular bone in the femoral head of strepsirrhine primates. Journal of Human Evolution, 43(1), 1–26.
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods, 9(7), 676–682.
Schneider, C. A., Rasband, W. S., & Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9(7), 671–675.
Acknowledgements
I thank the curator—Frieder Mayer—assistant curators—Christiane Funk, Steffen Bock, and Anna Rosemann—and preparator—Detlef Willborn—of the extant mammals collection of the Museum für Naturkunde Berlin (MfN), for giving access to the specimens under their care. I am also indebted to Johannes Müller, Kristin Mahlow, and Martin Kirchner (µCT lab, MfN) for providing facilities to acquire and analyse CT-scans and helping with data acquisition. I was funded by the German Research Council (Deutsche Forschungsgemeinschaft; Grant Number AM 517/1–1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Amson, E. Overall Bone Structure as Assessed by Slice-by-Slice Profile. Evol Biol 46, 343–348 (2019). https://doi.org/10.1007/s11692-019-09486-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-019-09486-6