Abstract
The function of natural killer (NK) cells after allogeneic hematopoietic stem cell transplantation (allo-HSCT) is regulated by the balance between inhibitory KIRs (iKIRs) and activating KIRs (aKIRs). However, few studies have examined the subsequent expression of KIR genes unique to the donor. We defined the set of KIR genes expressed only in the donor and designed a method for measuring the expression of these KIR genes by quantitative real-time polymerase chain reaction (RT-qPCR) based on genetic cloning techniques. In this study, we evaluated the recovery pattern of KIR genes in 252 donor-recipient pairs. The expression of each KIR unique to the donor was in line with that of KIR genes shared by the donor and recipient, such as KIR2DS1, KIR3DS1, KIR2DS4, or KIR2DS3. The timing of the peak mRNA expression of aKIRs unique to the donor was inconsistent but occurred within the first 3 months posttransplantation, whereas the peak mRNA expression of iKIRs was consistently observed in the third month after transplantation. The expression of KIR2DL2 in the third month posttransplantation was significantly higher in the transplant recipients than in the donors (p = 0.01). The KIR2DL1 and KIR3DL1 levels in the transplant recipients in the second and third months posttransplantation were also obviously higher than the donor levels (p < 0.0001). Thus, these observations should be considered when attempting to predict the correlation between mRNA expression and prognosis after allo-HSCT.
Similar content being viewed by others
Introduction
Natural killer (NK) cells, as a type of lymphocyte, might act early in the immune response against tumor cells, and the absolute number of NK cells approximately 4 weeks after transplantation might exceed the number found in healthy individuals (Terszowski et al. 2012). Ruggeri et al. (2002) proposed that donor-derived NK cells can potentially show alloreactivity that results in graft-versus-leukemia (GVL) effects without causing graft-versus-host disease (GVHD). The NK cell receptor repertoire is individual and mainly determined by the KIR genotype. Therefore, the function of NK cells is regulated by inhibitory and activating receptors for major histocompatibility complex (MHC) class I molecules (Farag et al. 2002). KIR gene clusters are divided into haplotypes A and B. Haplotype A mainly encodes iKIRs, with the exception of activating KIR (aKIR) KIR2DS4, whereas haplotype B displays greater variability and includes KIRs that are not present in haplotype A, such as KIR2DL2, 2DL5, 2DS1, 2DS2, 2DS3, 2DS5, and 3DS1 (Hsu et al. 2002).
We previously studied the distribution of specific donor-derived KIR genotypes after unrelated allogeneic hematopoietic stem cell transplantation (allo-HSCT) (Zhang et al. 2017). In recent years, the use of transplant donors possessing KIR haplotype B has been associated with a better long-term prognosis than the use of donors with KIR haplotype A (Bao et al. 2015; Cooley et al. 2014; Symons et al. 2010; Neuchel et al. 2017; Mehta et al. 2018). Elfishawi et al. (2017) reported that KIR2DS3 derived from donors can encourage rapid leukocyte engraftment. Using matched sibling donors, Stringaris et al. (2010) found that donor KIR2DS1 and KIR3DS1 are associated with reduced relapse in acute myeloid leukemia (AML) patients. We previously reported that donor KIR centromeric B-specific gene motifs exert a positive effect on the survival of standard risk AML/myelodysplastic syndrome (MDS) patients (Bao et al. 2015). Haplotype B includes more aKIR genes than haplotype A, and thus provides a basis for the expanding body of research on aKIRs unique to the donor and the corresponding prognoses. Hence, we separately quantified the mRNA levels of iKIRs and aKIRs by RT-qPCR and established reference intervals for each functional KIR gene in the donors and recipients at different timepoints. Our aim was to observe and compare the changes in the mRNA expression of KIR genes, particularly those found only in the donor, and evaluate their utility as prognostic markers after transplantation.
Materials and methods
Patients, donors, and clinical characteristics
Samples were collected from 252 donor-recipient pairs undergoing allo-HSCT between August 2011 and December 2018 at the First Affiliated Hospital of Soochow University (Table 1). The 252 enrolled patients consisted of 161 males and 91 females with a median age of 31 years (range, 8~63 years). The donors, who consisted of 174 males and 78 females with a median age of 31 years (range, 10~64 years), underwent physical examinations prior to transplantation. The patients had diseases that included AML (n = 109), acute lymphoblastic leukemia (ALL) (n = 69), chronic myelogenous leukemia (CML) (n = 21), MDS (n = 18), aplastic anemia (AA) (n = 15), and other diseases (n = 20).
Sample collection
Ten milliliters of peripheral blood (PB) or bone marrow (BM) was collected from all the donors on the day of transplantation. PB (2 ml) was obtained from all the recipients at 1-month intervals for the first 6 months after allo-HSCT. After this initial period, PB (2 ml) was obtained from the recipients every 3 months, due to the difficulty associated with follow-ups, and missing samples were mainly a result of mortality or disease progression. Sample collection was scheduled at the following times after transplantation: 28 ± 4 days (referred to as the 1 M samples), 58 ± 4 days (2 M), 88 ± 4 days (3 M), 118 ± 4 days (4 M), 148 ± 4 days (5 M), 178 ± 4 days (6 M), 210~270 days (7~9 M), and 300~360 days (10~12 M).
KIR genotyping
Genomic DNA was extracted from PBMCs using a Gentra Puregene Blood Kit (QIAGEN, USA). Genotyping was performed by PCR amplification with sequence-specific oligonucleotide probes (PCR-SSOP), using a KIR Genotyping SSOP Kit (One Lambda, USA). The PCR amplification reactions were performed using a Perkin Elmer GeneAmp 9700 thermal cycler (Perkin Elmer, USA). The data analyses were performed using a Luminex system (USA), and the results were compared with the “Allele Frequency Net Database” (http://www.allelefrequencies.net/kir6001a.asp) (Gonzalez-Galarza et al. 2015). KIR genotyping was performed for all of the enrolled recipient-donor pairs and repeated in the first month after transplantation, and the results revealed that the KIR genotypes of 105 recipients who received specific donor-derived KIR genes transformed into those of their donors during the first month. The distribution of donor-recipient KIR genotypes is listed in Table 1.
Experimental groups
According to the KIR genotype, a recipient with the same KIR genotype as the donor will not display specific donor-derived KIR genes, whereas a recipient who does not share the donor’s KIR genotype will show specific donor-derived KIR genes. Hence, we divided the 252 pairs into two groups: (1) a specific donor-derived KIR gene group (n = 105) and (2) a nonspecific donor-derived KIR gene group (n = 147). The most common combination of specific donor-derived KIR genes and the donor-recipient KIR genotypes can be found in Table 2.
Real-time reverse transcription–qPCR analysis
Total RNA samples from 252 donor-recipient pairs were isolated using the TRIzol Reagent (Invitrogen, USA). One microgram of total RNA was reverse-transcribed using 100 ng of random primers and M-MLV Reverse Transcriptase (Promega, USA). The cDNA templates for PCR were generated by reverse transcription from 2 μg/μl total RNA. Each amplification process involved an individual reaction and was performed in a volume of 25 μl, which included 0.5 μl of the forward primer, 0.5 μl of the reverse primer, 0.3 μl of the TaqMan MGB probe, 12.5 μl of the PCR mix, 7.2 μl of double-distilled water, and 4 μl of cDNA. The 50-ml manually formulated PCR mix contained 10 ml of 10 × buffer, 6 ml of Mg2+ (25 mM), 1 ml of AmpliTaq Gold DNA polymerase (ABI, USA), 0.1 ml of Uracil-DNA glycosylase (Shinegene, China), 1.2 ml of dUTP, 1.2 ml of dATP, 1.2 ml of dCTP, 1.2 ml of dGTP, 1.2 ml of dTTP (Sangon, China), and the rest double-distilled water. For the PCR reactions, the samples were heated at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. The specificity of the PCR assays was examined through the use of no-template control (NTC) and negative control (NC) samples. No amplification was found in the NTC or NC samples. The PCR assays for all KIR genes and the housekeeping gene ABL were optimized on a LightCycler 480 (Roche, Germany). In each amplification reaction, the correlation coefficient of the standard curve was greater than 0.99, and the slope of the curve was between − 3.3 and − 3.5. The primers and probes were designed with Primer Premier 5.0 (USA) and Primer Express 3.0 (USA), and were synthesized by Thermo (USA). All the sequences of the primers and probes are summarized in Table 3. The copy numbers of the KIR genes are expressed as KIR/ABL ratios and were calculated according to the following formula: (KIR/total ABL ratio × 10,000). This value was defined as the copy number of the KIR gene for every 10,000 ABL gene copies in all of the nucleated cells in the blood.
Statistical analysis
SPSS v20.0 (IBM, USA) was used for the statistical analyses, and graphs were generated using GraphPad Prism 5.0 (GraphPad Software, USA). The KIR gene copy numbers in the transplant recipients are expressed as medians (ranges) and were compared with those in the donors using the Mann-Whitney U test. The contribution of the sex of the recipients and donors was analyzed by a chi-squared test. Fisher’s exact test was utilized to compare the distribution of KIR genotypes among the groups. Continuous variables for AML, ALL, CML, MDS, AA, and other diseases were analyzed by one-way ANOVA.
p values < 0.05 were considered statistically significant, and two-sided tests were used for all the analyses.
Results
Recipient and donor clinical characteristics
Among the 252 enrolled patients, no significant differences were observed among the groups in terms of the age or gender of the recipients and donors. In contrast, significant differences in the total KIR genotype distribution in recipient-donor pairs were found among the groups (p = 0.039). For further cross-group comparisons, we performed a post hoc test with adjusted standardized residuals and considered residuals with an absolute value of more than three to indicate positive significant differences (Agresti 2002). In this manner, we found that the number of BX/AA samples was markedly lower than expected in CML, and higher than expected in the other groups, which included AML, ALL, MDS, and AA. Notably, the distribution of BX/AA donor-recipient pairs did not show significant differences among the AML, ALL, MDS, and AA groups (the absolute value of the residuals was not higher than three). No significant differences in the BX/BX recipient-donor category were observed (p = 0.56).
Analysis of KIR2DL1 and KIR3DL1 expression under the KIR/HLA mismatched model
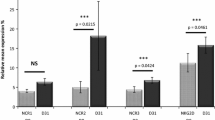
In the 252 transplantation pairs, 67 pairs exhibited matched KIR/HLA ligands and 185 pairs consisted of mismatched KIR/HLA ligands. According to whether the specific HLA ligands matched between the donors and recipients, we analyzed the expression of KIR2DL1 and KIR3DL1 in the donors and recipients during the first 3 months after transplantation. We found no significant difference in KIR2DL1 expression in the HLA C1/C1, HLA C1/C2 or HLA C2/C2 positive groups (Fig. 1). The same result was found for KIR3DL1 expression among the HLA Bw6/Bw6, HLA Bw4/Bw6 or Bw4/Bw4 positive groups (Fig. 2).
Division of specific donor-derived KIR genes and nonspecific donor-derived KIR genes
Based on the KIR genotyping results, we identified the specific donor-derived KIR genes in the aforementioned experimental group and recorded the high-frequency combinations of specific donor-derived KIR genes in the group (Table 2). The rest of the combinations were not described in detail due to their rarity and small sample sizes. For specific donor-derived KIR genes, the patterns of the donor and recipient KIR genotypes were diverse, and the most frequent combination was donor Bx1 vs recipient AA, followed by donor Bx2 vs recipient AA. In addition, among most of the donor AA vs recipient Bx pairs that were found in this study, specific donor-derived KIRs were not observed except in the case of donor AA vs recipient BB1 or BB6. As for nonspecific donor-derived KIR genes, the highest frequency pair was donor AA vs recipient AA (n = 78, 53.1%), followed by donor Bx1 vs recipient Bx1 (n = 19, 12.9%), and then donor Bx2 vs recipient Bx2 (n = 11, 7.5%).
Expression of KIR genes in the specific and nonspecific donor-derived groups
The median expression of KIR2DS1 in the specific donor-derived group exceeded in the donors in the first month after allo-HSCT and reached its peak value of 2372.2 (range, 817.5~6828.9) in the second month posttransplantation (Fig. 3a). However, the expression decreased after this peak until it approached the donor level at 1-year posttransplantation. Interestingly, the median expression of KIR2DS1 in the nonspecific donor-derived group was similar to that in the specific donor-derived group and reached its peak value of 2722.2 (range, 938.9~7865.9) in the second month posttransplantation. No significant differences in expression were found between these two groups at each time point.
The median expression of KIR2DS3 in the specific donor-derived group in the first month posttransplantation was 2082.3 (range, 198.9~4727.9) (Fig. 3b), and the expression gradually decreased until reaching a level that was approximately equal to the donor level within 1-year posttransplantation. In the nonspecific donor-derived group, the level observed in the first month posttransplantation was 1955.7 (range, 854.2~3434.3), and the observed trend was similar to that found in the specific donor-derived group. No significant differences were found between the two groups at any time point. The expression of KIR2DS3 in the nonspecific donor-derived group was not significantly different from that in the donors during the study period, whereas the expression in the specific donor-derived group in the second month posttransplantation was higher than that observed in the donors (1486.8 (range, 590.1~4351.0) vs 910.7 (range, 282.4~2580.8), p = 0.04)).
The median expression of KIR2DS4 in the two groups gradually increased until the third month posttransplantation. In the second month, the expression of KIR2DS4 in the specific donor-derived group was lower than that in the nonspecific donor-derived group (1736.0 (range, 1120.4~4085.2) vs 3027.78 (range, 913.3~7610.1), p = 0.12)) (Fig. 3c). The highest KIR2DS4 expression in the specific donor-derived group was 3409.1 (range, 1670.9~4090.9), which was lower than that in the nonspecific donor-derived group (3767.2 (range, 1198.3~8985.8)), but this difference was not significant.
The median expression of KIR3DS1 in the specific donor-derived group continuously increased during the study period, and in the third month, the median KIR3DS1 expression in the specific donor-derived group reached a value of 3809.2 (range, 313.4~7158.5) (Fig. 3d), which was higher than the expression level detected in the donors and recipients at all tested timepoints. During the rest of the study period, decreases in the KIR3DS1 expression levels with small intermittent fluctuations were observed in both the specific donor-derived and nonspecific donor-derived groups. No significant differences were observed between the two groups.
Comparison of iKIR and aKIR expression
The expression of iKIRs, including KIR2DL1, KIR2DL2, and KIR3DL1, continuously increased until the third month after transplantation, and significant differences in their expression, particularly that of KIR2DL1 and KIR3DL1 in the second and third months posttransplantation (p < 0.0001), were found compared with the donor level (Fig. 4a). In the third month, the highest expression was found for KIR3DL1 (2361.0 (range, 386.1~6729.8)), followed by KIR2DL2 and KIR2DL1, the latter of which exhibited a value of 947.7 (range, 205.2~3743.6). The expression of KIR2DL2 in the third month was clearly higher in the recipients than in the donors (1762.4 (range, 354.8~5919.3) vs 714.3 (range, 355.9~2991.3, p = 0.01)). In contrast, in the first month posttransplantation, the KIR gene levels in the recipients were not significantly different from those in the donors, although the median expression levels were higher than those in the donors. Starting in the third month, the frequencies of KIR2DL1, KIR3DL1, and KIR2DL2 decreased until they reached the donor levels 1 year after allo-HSCT. The same trends were observed for the reconstituted copy numbers.
The median expression levels of KIR2DS3 and KIR2DS5 reached their peak values in the first month posttransplantation, whereas KIR2DS1 expression reached its peak in the second month and the highest KIR2DS2, KIR2DS4, and KIR3DS1 expression levels were detected in the third month. The expression of the target KIR genes gradually increased to the peak value and then began to decrease, until reaching the donor level within 1 year after transplantation, and the same trend was found for all the reconstituted KIR genes that were studied. The copy numbers of most aKIRs in the recipients were not significantly different from those in the donors; however, the median expression of KIR2DS3 in the recipients in the second month was higher than that in the donors (1486.8 (range, 590.1~4351.0) vs 910.7 (range, 282.4~2580.8), p = 0.04)) (Fig. 4b), and the median expression of KIR2DS2 in the recipients in the second month was higher than that in the donors (1169.7 (range, 435~3596.7) vs 640.7 (range, 430.2~1723.4), p = 0.0043)) (Fig. 4b).
The copy numbers of iKIRs were different from those of specific donor-derived aKIRs. The expression of iKIRs remained markedly elevated in the recipients between the first and third months after transplantation, whereas the expression of aKIRs in the recipients was higher than that in the donors by only a nonsignificant margin. The peak values for KIR2DL1, KIR3DL1, and KIR2DL2 were detected in the third month posttransplantation, whereas the peak KIR2DS3 and KIR2DS5 copy numbers were obtained in the first month, and the peak value for KIR2DS1 expression was reached in the second month. However, the expression of both iKIRs and aKIRs reached the donor levels within the first month after transplantation gradually increased to reach their peak values and then decreased to return the donor levels within 1 year.
Discussion
NK cells integrate signals triggered by interaction of target cell ligands with an array of activating and inhibitory KIRs after allo-HSCT. Specific donor-derived KIR genes can be distinguished from nonspecific donor-derived ones through KIR genotyping of donor-recipient pairs. In this study, we observed that 105 of the 252 enrolled donor-recipient pairs possessed specific donor-derived KIR genes. It was a shortcoming that no detection method for the expression of specific donor-derived KIR genes was performed in the previous studies. One of the most common methods used to measure the expression of KIR genes is flow cytometric analysis, which can hardly distinguish the expression of aKIRs and iKIRs, let alone that of specific donor-derived KIR genes. We previously dynamically detected the expression of KIR2DL1/3DL1 protein and mRNA in NK cells (Hu et al. 2017). The median protein expression level of KIR3DL1 in the posttransplant recipients in the first month exceeded the donor level and reached the highest value in the third month, and this finding potentially implicates the reconstitution of NK cells in the expression of membrane protein. The comparison of the recovery trend of KIR3DL1 protein and mRNA revealed that the mRNA expression of KIR3DL1 was in line with that of the membrane protein, even though the recovery of KIR2DL1 mRNA expression occurred earlier than that of the protein, and this delay might be due to the interaction between KIR2DL1 and HLA class I ligands and the polymorphisms in different KIR repertoires. Giebel et al. (2010) evaluated the recovery of the expression of NK cell iKIRs through cytometric analysis. Gallez-Hawkins et al. (2011) studied the mRNA expression of two aKIRs, KIR2DS4 and KIR2DS2, during the postinfection phase, and various novel KIR genotype methods have been used to study polymorphisms of KIR genes (Chaisri et al. 2018; Kitpoka et al. 2016; Wang et al. 2012; Yao et al. 2019). Chen et al. (2009) designed a quantitative KIR RNA type assay and found leukemia patients exhibited lower KIR expression before transplantation than healthy donors. These researchers also hypothesized that mobilization might transiently enhance donor KIR expression but does not affect the balance between iKIRs and aKIRs. Denis et al. (2005) found that the delayed appearance of KIR transcripts posttransplantation might be associated with lower absolute numbers of CD45+, CD3−, CD16+, CD56+ NK cells and a higher risk of acute GVHD. These results suggest that KIR expression undergoes kinetic changes that might influence the process and outcome of transplantation. However, our predictions call for an innovative method with better quantification and dynamics than other previously developed tests. Hence, we designed a method for measuring the mRNA expression of KIR genes based on genetic cloning techniques and applied for the domestic patents on our method. The RT-qPCR assays for all the KIR genes and the ABL housekeeping genes were optimized simultaneously because ABL is expressed at similar levels in healthy individuals and patients (Beillard et al. 2003). Considering the potential effect of KIR/HLA mismatch on mRNA expression, we analyzed the mRNA expression of KIR2DL1 and KIR3DL1, and found no significant difference due to KIR/HLA mismatch. Hence, the mRNA expression patterns obtained in our study could be used for distinguishing specific and nonspecific donor-derived KIR genes. For instance, a donor Bx1 vs a recipient AA, specific donor-derived KIR genes allowed for detecting mRNA expression can be regard as KIR3DS1, KIR2DL5, KIR2DS5, and KIR2DS1. According to our research, the expression of each specific donor-derived KIR gene was in line with that of the nonspecific donor-derived KIR gene. Therefore, we also evaluated the recovery pattern of the expression of aKIRs and iKIRs at different timepoints after transplantation.
The specific donor-derived KIR genes were mostly aKIRs, and only a small proportion were iKIRs. KIR3DL1 and KIR2DL1 account for the majority of specific donor-derived iKIRs. The ligands of these two proteins have been thoroughly studied and discussed, yet the research on that of aKIRs was still unknown (Anfossi et al. 2006; Boudreau et al. 2016; Fernandez et al. 2005; Heidenreich and Kroger 2017; Kim et al. 2005; Yawata et al. 2008). The main specific donor-derived aKIRs are KIR2DS1, KIR2DS3, KIR2DS5, and KIR3DS1. Based on the findings obtained in this study, we established the chronology of the peak values of aKIRs after transplantation. Specifically, we found that the expression of KIR2DS3 and KIR2DS5 peaked in the first month after transplantation, and KIR2DS1 expression reached its highest level in the second month, and that the highest values for KIR2DS4 and KIR3DS1 expression were detected in the third month. Moreover, each aKIR showed a different expression level at every time point after transplantation, and their expression was dynamic but relatively stable. Therefore, the aKIRs were derived from the donors and could be used for comparison with pretransplantation results and for clinical follow-up. The correlation between mRNA expression changes, and manifestations are being analyzed in our ongoing work. In addition, the expression of specific donor-derived KIR2DS3 and KIR2DS2 in the second month posttransplantation was significantly different from the donor levels. These results were likely obtained due to the small sample size and the inequitable genotype distribution of KIR2DS2 in the Chinese Han population. More specific samples are necessary to characterize the expression level in detail. Compared with the aKIR repertoires, iKIRs involve not only definite ligands but also different arrangements in the genomic structure of KIRs (Barten et al. 2001; Martin et al. 2000, 2004). We found that the iKIR repertoires exhibited a different recovery trend from the aKIRs, and their trend involved a more rapid development and higher expression level posttransplantation relative to the donor level. Interestingly, all the examined iKIRs reached their highest expression levels in the third month posttransplantation.
Rapid recovery indicates a significant role for NK cells in the early phase after transplantation. It is well known that allo-HSCT faces two difficulties that require breakthroughs, i.e., the restoration of hematopoiesis and the reconstitution of the immune system. In this study, we focused on immune reconstitution and designed a method for quantitatively measuring the expression of specific donor-derived KIR genes. The dynamic changes in the expression of these KIR genes could serve as a biomarker for predicting and evaluating NK cell functional recovery after transplantation. As previous study indicated that a beneficial alloreactive NK cell response can be induced in recipients who lack a KIR ligand for donor-derived educated KIRs (Cooley et al. 2018). Several studies have hypothesized that a missing HLA ligand for donor inhibitory KIRs is associated with no-longer-suppressed inhibitory signals, enhanced cytotoxicity, and increased affinity between activating receptors and their ligands. We are studying, whether under discrepant KIR/HLA models, the levels of KIR mRNA expression exert an influence on the occurrence of GVHD, overall survival, and relapse-free survival in patients with malignant hematologic diseases.
References
Agresti A (2002) Categorical data analysis, vol 2002, 2nd edn. Wiley, New York, p 81
Anfossi N, André P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagné F, Ugolini S, Vivier E (2006) Human NK cell education by inhibitory receptors for MHC class I. Immunity 25:331–342. https://doi.org/10.1016/j.immuni.2006.06.013
Bao XJ, Wang M, Zhou HF, Zhang H, Wu X, Yuan X, Li Y, Wu D, He J (2015) Donor killer immunoglobulin-like receptor profile Bx1 imparts a negative effect and centromeric B-specific gene motifs render a positive effect on standard-risk acute myeloid leukemia/myelodysplastic syndrome patient survival after unrelated donor hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 22:232–239. https://doi.org/10.1016/j.bbmt.2015.09.007
Barten R, Torkar M, Haude A, Trowsdale J, Wilson MJ (2001) Divergent and convergent evolution of NK-cell receptors. Trends Immunol 22:52–57
Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, Delabesse E, Macintyre E, Gottardi E, Saglio G, Watzinger F, Lion T, van Dongen J, Hokland P, Gabert J (2003) Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using 'real-time' quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR) - a Europe against cancer program. Leukemia 17:2474–2486. https://doi.org/10.1038/sj.leu.2403136
Boudreau JE, Liu XR, Zhao Z, Zhang A, Shultz LD, Greiner DL, Dupont B, Hsu KC (2016) Cell-extrinsic MHC class I molecule engagement augments human NK cell education programmed by cell-intrinsic MHC class I. Immunity 45:280–291. https://doi.org/10.1016/j.immuni.2016.07.005
Chaisri S, Traherne JA, Jayaraman J, Romphruk A, Trowsdale J, Leelayuwat C (2018) Novel KIR genotypes and gene copy number variations in northeastern Thais. Immunology 153:380–386. https://doi.org/10.1111/imm.12847
Chen X, Knowles J, Barfield RC, Kasow KA, Madden R, Woodard P, Srivastava DK, Horwitz EM, Handgretinger R, Hale GA (2009) A novel approach for quantification of KIR expression in healthy donors and pediatric recipients of hematopoietic SCTs. Bone Marrow Transplant 43:525–532. https://doi.org/10.1038/bmt.2008.352
Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SG, Spellman S, Haagenson MD, Saeturn K, Ladner M, Trachtenberg E, Parham P, Miller JS (2014) Donor killer cell Ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol 192:4592–4600. https://doi.org/10.4049/jimmunol.1302517
Cooley S, Parham P, Miller JS (2018) Strategies to activate NK cells to prevent relapse and induce remission following hematopoietic stem cell transplantation. Blood:blood-2017-08-752170. https://doi.org/10.1182/blood-2017-08-752170
Denis L, Gagne K, Gueglio B, Kerdudou N, Milpied N, Simon P, Follea G, Bonneville M, Harousseau JL, Bignon JD (2005) NK-KIR transcript kinetics correlate with acute graft-versus-host disease occurrence after allogeneic bone marrow transplantation. Hum Immunol 66:447–459. https://doi.org/10.1016/j.humimm.2005.01.008
Elfishawi SM, Mossallam GI, El-Fattah RA, El-Haddad A, Kamel AM (2017) The effect of killer cell immunoglobulin-like receptor genotype on outcome of hematopoietic stem cell transplantation from matched sibling. Hum Immunol 78:684–691. https://doi.org/10.1016/j.humimm.2017.10.004
Farag SS, Fehniger TA, Ruggeri L, Velardi A, Caligiuri MA (2002) Natural killer cell receptors: new biology and insights into the graft-versus-leukemia effect. Blood 100:1935–1947. https://doi.org/10.1182/blood-2002-02-0350
Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH (2005) A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood 105:4416–4423. https://doi.org/10.1182/blood-2004-08-3156
Gallez-Hawkins GM, Franck AE, Li X, Thao L, Oki A, Gendzekhadze K, Dagis A, Palmer J, Nakamura R, Forman SJ, Senitzer D, Zaia JA (2011) Expression of activating KIR2DS2 and KIR2DS4 genes after hematopoietic cell transplantation: relevance to cytomegalovirus infection. Biol Blood Marrow Transplant 17:1662–1672. https://doi.org/10.1016/j.bbmt.2011.04.008
Giebel S, Dziaczkowska J, Czerw T, Wojnar J, Krawczyk-Kulis M, Nowak I, Holowiecka A, Segatti A, Kyrcz-Krzemien S, Kusnierczyk P, Holowiecki J (2010) Sequential recovery of NK cell receptor repertoire after allogeneic hematopoietic SCT. Bone Marrow Transplant 45:1022–1030. https://doi.org/10.1038/bmt.2009.384
Gonzalez-Galarza FF, Takeshita LY, Santos EJ et al (2015) Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res 43:D784–D788. https://doi.org/10.1093/nar/gku1166
Heidenreich S, Kroger N (2017) Reduction of relapse after unrelated donor stem cell transplantation by KIR-based graft selection. Front Immunol 8:41. https://doi.org/10.3389/fimmu.2017.00041
Hsu KC, Chida S, Geraghty DE, Dupont B (2002) The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev 190:40–52. https://doi.org/10.1034/j.1600-065x.2002.19004.x
Hu X, He J, Zhang HH et al (2017) Immune reconstruct regularity profile of KIR2DL1 and KIR3DL1 in unrelated-donor allogeneic hematopoietic stem cell transplantation. Chin J Hematol 38:667–672. https://doi.org/10.3760/cma.j.issn.0253-2727.2017.08.004
Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, Yokoyama WM (2005) Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 436:709–713. https://doi.org/10.1038/nature03847
Kitpoka P, Tammakorn C, Chaisri S, Leelayuwat C, Mongkolsuk T, Thammanichanond D (2016) Genetic profiles of killer-cell immunoglobulin-like receptors and HLA ligands in Thai blood donors. Hum Immunol 77:470–475. https://doi.org/10.1016/j.humimm.2016.04.019
Martin AM, Freitas EM, Witt CS, Christiansen FT (2000) The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics 51:268–280. https://doi.org/10.1007/s002510050620
Martin AM, Kulski JK, Gaudieri S, Witt CS, Freitas EM, Trowsdale J, Christiansen FT (2004) Comparative genomic analysis, diversity and evolution of two KIR haplotypes a and B. Gene 335:121–131. https://doi.org/10.1016/j.gene.2004.03.018
Mehta RS, Randolph B, Daher M, Rezvani K (2018) NK cell therapy for hematologic malignancies. Int J Hematol 107:262–270. https://doi.org/10.1007/s12185-018-2407-5
Neuchel C, Furst D, Niederwieser D et al (2017) Impact of donor activating KIR genes on HSCT outcome in C1-ligand negative myeloid disease patients transplanted with unrelated donors-a retrospective study. PLoS One 12:e0169512. https://doi.org/10.1371/journal.pone.0169512
Ruggeri L, Capanni M, Urbani E et al (2002) Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 295:2097–2100. https://doi.org/10.1126/science.1068440
Stringaris K, Adams S, Uribe M, Eniafe R, Wu CO, Savani BN, Barrett AJ (2010) Donor KIR genes 2DL5A, 2DS1 and 3DS1 are associated with a reduced rate of leukemia relapse after HLA-identical sibling stem cell transplantation for acute myeloid leukemia but not other hematologic malignancies. Biol Blood Marrow Transplant 16:1257–1264. https://doi.org/10.1016/j.bbmt.2010.03.004
Symons HJ, Leffell MS, Rossiter ND, Zahurak M, Jones RJ, Fuchs EJ (2010) Improved survival with inhibitory killer immunoglobulin receptor (KIR) gene mismatches and KIR haplotype B donors after nonmyeloablative, HLA-haploidentical bone marrow transplantation. Biol Blood Marrow Transplant 16:533–542. https://doi.org/10.1016/j.bbmt.2009.11.022
Terszowski G, Passweg JR, Stern M (2012) Natural killer cell immunity after transplantation. Swiss Med Wkly 142:w13700. https://doi.org/10.4414/smw.2012.13700
Wang HD, Zhang FX, Shen CM, Wu YM, Lv YG, Xie ST, Yang G, Qin HX, Fan SL, Zhu BF (2012) The distribution of genetic diversity of KIR genes in the Chinese Mongolian population. Hum Immunol 73:1031–1038. https://doi.org/10.1016/j.humimm.2012.07.317
Yao Y, Shi L, Yu J, Liu S, Tao Y, Shi L (2019) Distribution of killer-cell immunoglobulin-like receptor genes and combinations of their human leucocyte antigen ligands in 11 ethnic populations in China. Cells 8. https://doi.org/10.3390/cells8070711
Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P (2008) MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood 112:2369–2380. https://doi.org/10.1182/blood-2008-03-143727
Zhang HH, He J, Bao XJ et al (2017) Distribution of donor-specific aKIR after unrelated allogeneic hematopoietic stem cell transplantation. Chin J Hematol 38:421–426. https://doi.org/10.3760/cma.j.issn.0253-2727.2017.05.013
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81671549), the Jiangsu Province Medical Innovation Team (CXTDB2017009), the Jiangsu Provincial Key Research and Development Program (BE2019656), and the Collaborative Innovation Center of Hematology (SX21100117).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This protocol was approved by the local ethics committee. In accordance with the ethical guidelines of the Declaration of Helsinki; written informed consent was obtained from all the subjects.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, Y., Wang, T., Hu, X. et al. Study of KIR gene expression at the mRNA level in specific donor-derived NK cells after allogeneic HSCT. Immunogenetics 72, 135–141 (2020). https://doi.org/10.1007/s00251-019-01153-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-019-01153-6