Abstract
The phylogeography and possible origins of the moss Rhytidium rugosum (Hedw.) Kindb. in the European Alps are studied based on information from the nuclear internal transcribed spacers 1 and 2 and a portion of the gene region for glyceraldehyde 3-phosphate dehydrogenase for 364 European specimens. Seventy-three Alps specimens were sampled from W Switzerland to W Austria, and were divided into four regional populations along a West-South-West (WSW) to East-North-East (ENE) gradient. These populations were compared with similar ones previously studied in other parts of Europe. The ENE-most Alps population, located ENE of the Adige break zone, deviates genetically from the other three. The two WSW-most populations of the Alps appear to be relatively isolated from most of the European populations outside the Alps, whereas the two ENE ones are similar to populations of northern Scandinavia. Populations in between the Alps and the Scandinavian mountain range deviate from those to the north and south, possibly due to low effective population sizes, earlier bottleneck events, or colonization from different source populations. Haplotype diversity and number of private haplotypes are marginally higher in the Alps than in Scandinavia. It is suggested that European Rhytidium originated from late glacial maximum refugia in (1) E-NE Europe, (2) in between the Late Glacial Maximum ice shields of Scandinavia and the Alps, and (3) S, SW, and W of the Swiss Alps. Those of the E Alps potentially originated mainly in E-NE Europe and those of the W Alps in the S, SW, and W.
Similar content being viewed by others
Introduction
Numerous investigations have focused on the post-glacial colonization or phylogeographic patterns of different portions of Europe, using molecular evidence (e.g. Brochmann et al. 2003; Hedenäs 2015; Hewitt 2000; Jaarola et al. 1999; Kyrkjeeide et al. 2014; Parducci et al. 2012; Taberlet et al. 1998). Several recent studies concerned plants, especially flowering plants, in the European Alps (from here on called ‘the Alps’) (e.g. Gugerli and Holderegger 2001; Schönswetter et al. 2005; Thiel-Egenter et al. 2011). Source populations for late- to post-glacial colonization of the Alps by plants occurred in glacial refugia in the lowlands, often along the margins of the glaciated area, or on nunataks within the ice shield, and the molecular evidence for such refugia was summarized by Schönswetter et al. (2005). Many flowering plant species of the Alps seem not yet to have colonized all potential habitats due to limited dispersal abilities (Dullinger et al. 2012; Schönswetter et al. 2005). Where environmental conditions hinder efficient dispersal of species or intraspecific entities of different origins, break zones are found where the turnover in species and/or genotypes within species is markedly higher than elsewhere (Thiel-Egenter et al. 2011). More break zones exist at the species compared with the genotype level among flowering plants, due to differences in dispersal potential between species (dispersed by seeds) and genotypes (by pollen and seeds). Two major break zones where species and genotype patterns coincide occur around the Aoste valley in the west and around the Adige valley in the east (Thiel-Egenter et al. 2011).
In organisms like bryophytes and fungi, which disperse by spores, sizes of dispersal units are usually in the lower range of pollen grains. This means that the dispersal potentials of both species and intraspecific entities are similar to that of pollen in flowering plants. Spores below c. 20 µm are efficiently dispersed by wind (Wilkinson et al. 2012) and in open, windy environments with few or no trees also small vegetative fragments are easily spread (Flø and Hågvar 2013; Miller and Ambrose 1976). Phylogeographic patterns of such organisms should thus be more similar to those proposed for small organisms in a meta-community perspective (De Meester 2011) than to the average flowering plant species. Factors that could contribute to intraspecific structure in spore-dispersed organisms include past or present dispersal barriers, the ‘founder takes all’ density-dependent principle (Waters et al. 2013), niche specialization (Buckley et al. 2013; Mikulášková et al. 2014), and genetic drift in isolated populations (Frankham et al. 2002). Based on these characteristics and processes, we can hypothesize that break zones in spore-dispersed organisms, both at the species and intraspecific levels, should likely mirror those of genotypes within widespread vascular plants.
The number of studies of spore-dispersed organisms that cover large portions of the Alps and its surroundings with sufficient sampling density to detect Alps-wide patterns is limited. Extensive work exists for the lichenized fungus Lobaria pulmonaria (L.) Hoffm. and suggests immigration from glacial refugia in Southern Italy-Balkan, and potentially from additional refugia for the fungal component (Widmer et al. 2012). Within the Alps, the presence of sexual reproduction in a specific L. pulmonaria population appears important to explain its genetic diversity (Zoller et al. 1999). Further, these authors found that geographical isolation may result in clear genetic differences between populations, whereas no overall pattern congruent with the break zones identified for vascular plants was found. This is likely explained by the geographic sampling of Zoller et al. (1999), who had a conservation genetic rather that phylogeographic focus (see also Walser et al. 2005). The sampling density for bryophytes in studies where the Alps were included was either too low to detect Alps-wide patterns (e.g. Hedenäs 2010, 2012; Hedenäs and Eldenäs 2007) or only a limited region of the Alps was covered in detail (Kophimai 2013).
The present investigation explores the intraspecific molecular variation of the moss Rhytidium rugosum (Hedw.) Kindb. in the Alps to see whether its intraspecific geographical variation is consistent with what was hypothesized above for spore-dispersed plants and to possibly trace its post-glacial origin. In Rhytidium, the variation in the nuclear internal transcribed spacers 1 and 2 (ITS) and a portion of the gene region for glyceraldehyde 3-phosphate dehydrogenase (gpd) are considerable and shows a geographic pattern largely consistent with what is known about the late- to post-glacial history of Scandinavia (Hedenäs 2014, 2015). Compared with Scandinavian regions, Hedenäs (2014) found a high haplotype diversity in a Rhytidium population in Val Poschiavo in SE Switzerland. It was suggested that the more complicated patterns of glacial refugia in and close to the Alps (Schönswetter et al. 2005) than in the almost completely ice-covered Scandinavia (Lambeck et al. 2010; Westergaard et al. 2011; Vorren et al. 2013) during the Late Glacial Maximum (LGM, c. 30–16 kA BP) could explain this high diversity. Some European Rhytidium haplotypes that are unique to the Alps were found by Hedenäs (2015). However, such geographically restricted haplotypes were not more common in central Europe than in Scandinavia, as would have been expected if a larger proportion of the species’ total diversity had survived in the southern refugia than in the north (cf., Koch et al. 2006; Schönswetter et al. 2003). However, if there are a large number of haplotypes that are unique to the Alps, many of these likely went undetected in the earlier sparse sampling of this particular area (Hedenäs 2015). Whether the species immigrated to an area on a broad front or not could explain diversity differences (Alsos et al. 2009), but we should also expect a higher diversity in the Alps than in Scandinavia if more Rhytidium haplotypes survived the last glaciation in situ, in local glacial refugia, or if it immigrated from a larger number of lowland glacial refugia. Based on subfossil macro-remains, it can be inferred that Rhytidium was widely distributed in the European lowlands north of the Alps during the glacial period, and it still survives in interglacial refugia in some such areas with relaxed competition from flowering plants (Hedenäs 2014, 2015; Meinunger and Schröder 2007). Finally, lowlands where source populations for the extant occurrences likely occurred are, relative to the glaciated areas, both larger and more closely situated for the Alps than for most of Scandinavia.
To detect Alps-wide patterns of diversity and intraspecific structure, and to make comparisons with populations just to the north and northeast of the Alps, as well as Scandinavian and north-eastern European populations of Rhytidium, the present study samples the Alps much more densely than Hedenäs (2015). The following questions are addressed: (1) Do the Alps populations of Rhytidium show a geographic structuring similar to that in vascular plants? (2) Do haplotype and nucleotide patterns of the Alps and other regional populations of Rhytidium suggest colonization from different sources? (3) Is the intraspecific diversity of Rhytidium in the Alps in general higher than in Scandinavia, and is there a surplus of private haplotypes in the Alps compared with Scandinavia?
Materials and methods
Study species and material
Rhytidium rugosum is a large pleurocarpous moss, which general morphology was described by Hedenäs (2014). It is widespread in Europe, even if it is absent from large areas (Hedenäs 2015), in North America and Asia, and has scattered occurrences at high elevations in Central America and North and East Africa (Ochyra and Szmajda 1983). The species has separate male and female plants (dioicous) and sporophytes are nowadays rarely seen in Europe except in the Gudbrandsdalen area in southern Norway, where they are common and regularly produced (Hedenäs 2014, 2015). The moss produces spores, which are 10.5–15.0 µm in diameter and can disperse far by wind, and vegetative fragments that vary from a few mm to a few cm (Pfeiffer et al. 2006). The smaller such fragments can disperse relatively far in windy environments.
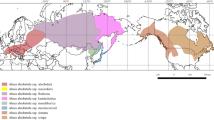
For Europe, 364 specimens (including one from Turkey) with complete sequence data are included. Ninety-nine of the specimens come from the Alps and Central Europe, including 58 specimens from the Alps for which sequences were generated in this study. Ten extra-European specimens from different portions of the range of the species are included in some of the overall analyses to place European Rhytidium in a global context. Three hundred thirty-eight of the specimens were grouped in eighteen regional populations based on geography, delimited as in Table 1, where the population acronyms are explained (cf., Figure 1). Populations A–D are contiguously located along a WSW-ENE gradient in the Alps, with 16–19 specimens per population. The other regional populations are located in Scandinavia (11 populations; 14–28 specimens in each), in the lowlands to the north and lowland to montane area northeast of the Alps (2; 13–14), and in NE Europe and NW-most Asia (1; 25). Further subdivision of the last area into geographical regions of sizes comparable to the other regions was prohibited by difficulties to get access to sufficient numbers of specimens. The earlier sampled 41 specimens from the Alps and Central Europe (Hedenäs 2014, 2015) belong to the regional populations as listed in Online Resource Table S1.
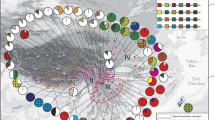
Map of the distribution of European and NW Asiatic haplotypes (cf., Online Resource Table S3) based on the nuclear ITS and gpd together for 364 Rhytidium rugosum specimens. The focal populations, which acronyms for the populations are explained in Table 1, include 338 of these specimens. The haplotypes are arranged clockwise according to their numbers. (The haplotypes are indicated in colour in the online version of the paper)
Potential differences between southern and northern Rhytidium populations of the Alps are explored by merging the three western regional populations, dividing this total set into a southern and northern component (indicated in Fig. 2), and by comparing these with each other and the lowland population to the north of these (N in Table 1). The southern component consists of samples from northern Italy and southern Switzerland (S Graubünden, Ticino, Valais) and the northern component includes the other Swiss samples. The easternmost population of the Alps is located E of the Adige break zone mentioned above and was excluded from this comparison to avoid introducing any bias due to potential differences between populations on the two sides of this break zone.
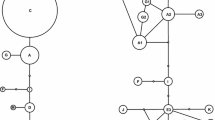
Longitudinal and latitudinal distribution of haplotypes (a; cf. Online Resource Table S3), and members of NeighborNet split network branches/groups (b; cf. Figure 3), based on ITS and gpd together, among the four regional populations A–D of the European Alps. The populations, explained in Table 1, include 19 (A), 19 (B), 18 (C), and 16 (D) specimens, respectively. The dotted line indicates the alternative division of the three western regional populations into a northern and southern component (see text)
Specimen data are provided in Hedenäs (2014, 2015) and Online Resource Table S2.
Molecular methods
The molecular methods used here are the same as in the previous study (Hedenäs 2015), and the reader is referred to this for details. In the present study, only the primers’ITS4bryo’ and ‘ITS5bryo’ (Stech 1999) were used for ITS, and ‘GPD-bryoF’ and ‘GPD-Rhy.rug-nestedR’ (Hedenäs 2014) for gpd.
Sequence editing and analysis
Nucleotide sequence fragments were edited and assembled for each DNA region using PhyDE® 0.9971 (http://www.phyde.de/index.html; accessed 6 January 2017). The assembled sequences were aligned manually in PhyDE®. Regions of partially incomplete data in the beginning and end of the sequences were identified, and were excluded from subsequent analyses. The gaps, when coded as present or absent, provided additional evidence to distinguish haplotypes and the analyses were performed with the insertions and deletions coded as single informative characters independent of their length. The sequence alignments used in the analyses are available on request. GenBank or European Nucleotide Archive (EMBL-ENA) accession numbers are listed in Hedenäs (2014, 2015) and Online Resource Table S2, respectively.
Paralogous ITS haplotypes are rarely encountered in bryophytes (but see Košnar et al. 2012). The ITS chromatograms generated in this study did not show ‘messy’ patterns or noise that could suggest paralogy, and the 5.8S gene was invariable among all samples (cf., Feliner and Rosselló 2007; Shaw et al. 2002). The revealed limited ITS variation was thus interpreted as being among homologous haplotypes. The variable portions of ITS are non-coding, whereas gpd includes both coding and non-coding elements, with considerable variation in both. Glyceraldehyde 3-phosphate dehydrogenase (gpd) is involved in many basic cell functions, such as the glycolysis (Guo et al. 2014; Sirover 1999), and selection related to physiological climate adaptation could thus possibly exist. However, preliminary separate analyses of the two markers did not suggest that certain gpd haplotypes are restricted to specific climatic regions, and the two markers are therefore analysed together.
The program TCS (Clement et al. 2000) was used to find which haplotypes exist based on ITS plus gpd, and for a preliminary evaluation of relationships among specimens in a haplotype context. Reticulation was revealed and, therefore, the statistical support for potential recombination was tested by the Φ w statistic (Bruen et al. 2006) as implemented in SplitsTree 4.12.6 (Huson and Bryant 2006). Because abundant reticulation occurred a split network was computed with the NeighborNet (NN) method as implemented in SplitsTree to visualize similarities or relationships among samples.
The null hypothesis in the following analyses is that there exist no differences in haplotype and nucleotide diversity or composition among the studied populations. To investigate patterns of haplotype variation among regional populations (cf., Table 1; Fig. 1), or between the N and S sides of the Alps, analyses of molecular variance (AMOVA) were performed with GENALEX 6.501 (Peakall and Smouse 2006, 2012). Pairwise Φ PT (an analogue of F ST, i.e. genetic diversity among populations) was estimated with GENALEX, and the same program was used to calculate the effective number of haplotypes (Ne) and the haplotype diversity (H) for each population. Arlequin ver. 3.5.1.3 (Excoffier and Lischer 2010) was used to calculate nucleotide diversity (π) and average number of pairwise differences among populations. To visualize the variation among all European regional populations based on pairwise Φ PT values and average numbers of pairwise nucleotide differences, Principal Coordinate Analyses (PCoA) were performed in GENALEX.
Geographical distances (km) between the mid-points of the eighteen European regions were estimated from the ‘Movable Type Scripts’ homepage for calculations of distance, bearing and more between latitude/longitude points (http://www.movable-type.co.uk/scripts/latlong.html; accessed 24 November 2016). This site uses the ‘haversine’ formula to calculate the great-circle distance between two points. The relation between inter-regional Φ PT or average numbers of pairwise nucleotide differences and inter-region distance was then evaluated by Mantel tests in GENALEX for all European regions. To decide whether the populations are stable in size, expanding, or decreasing, Tajima’s D test of selective neutrality was employed (Tajima 1989), using Arlequin. Tajima’s D test was preferred over Fu’s F S test (Fu 1997), because recombination levels are unknown (Ramírez-Soriano et al. 2008).
The geographical distribution of individual haplotypes was mapped for regional populations as well as for sampled European localities outside these populations (Fig. 1), to find potential connections with Rhytidium from outside the Alps. To compare the total numbers of haplotypes and private haplotypes in the Alps with the corresponding numbers in Scandinavia, the number of Scandinavian samples was adjusted to 73, the total number of samples from the Alps when one Austrian sample outside the four regional populations is included. This was done by randomly resampling 73 specimens 50 times from among the 229 Scandinavian specimens. Random numbers were generated at the site ‘random.org’ (https://www.random.org/; accessed 6 January 2017). The mean numbers of Scandinavian haplotypes and private haplotypes, with 95% confidence intervals, were used to compare with the numbers from the Alps.
Results
The total number of aligned ITS sites in the 374 studied Rhytidium rugosum specimens, after deletion of regions at the beginnings and ends that were incomplete for some specimens, was 720, including 8 sites with base substitutions and 2 coded indels. For gpd, the corresponding values were 797, with 22 base substitutions and 4 indels. The number of parsimony informative sites including indels was 6 for ITS and 18 for gpd. For ITS, the sequence length was 716–718 and for gpd 793–796. One additional ITS substitution was found in a British specimen included by Hedenäs (2015), but this specimen lacks gpd information and is not included here. The gpd sequence length is shorter than in Hedenäs (2015) since the used length was adapted to the slightly shorter portion of this marker that was retrieved for specimens from the Alps. However, all previously found variable sites are included.
In total, 55 haplotypes were found in the TCS analysis based on ITS and gpd (cf., Online Resource Table S3). Fifty of these occur in Europe and NW Asia (Fig. 1) and 23 in the Alps (Fig. 2a). No statistical support for recombination was found (p = 0.111; no indel coding included). The NeighborNet (NN) split network based on ITS and gpd together is shown in Fig. 3, with geographical origins of the samples indicated. Specimens from the regional populations of the Alps occur, with varying frequencies, in all the nine main NN branches or groups, whereas specimens from the neighbouring N and NE populations occur in four or five. The distribution of representatives of different NN branches/groups differs among the four populations of the Alps (Fig. 2b). For example, samples from NN branches ‘g’ and ‘i’ are most common in the two western regional populations, those of ‘f’ are most frequent in the east, and those of ‘j’ in the two middle populations.
NeighborNet split network for the 374 Rhytidium rugosum specimens with complete data, based on the combined nuclear ITS and gpd. The geographical distribution of samples from Europe and NW-most Asia are indicated in pie diagrams for the different major branches/groups of the network, which are indicated by the letters a–k. The size of a pie diagram corresponds with the number of European and NW Asian specimens in a group. Acronyms for the regional populations are indicated in the upper left box and are explained in Table 1 and Fig. 1, except for NEU (N European samples outside regional populations) and CEU (other European samples outside regional populations). In the pie diagrams, the geographical regions are always depicted in the order indicated by the arrow. Haplotypes of a branch/group are indicated by numbers, which correspond with those in Online Resource Table S3. Haplotypes in parentheses were found only outside the study area
Different haplotypes dominate different portions of Europe (Fig. 1). The number of haplotypes, number of private haplotypes, effective number of haplotypes (Ne), haplotype diversity (H), nucleotide diversity (π), and Tajima’s D are compared for the 18 regional populations in Table 2. Compared with other regional Rhytidium populations, all four populations of the Alps have high Ne, H, and π values. The same is true for the population to the northeast of the Alps (NE), whereas somewhat lower values are found for the lowland population to the north (N). The values of population N are nevertheless higher than in several Scandinavian populations with clearly lower values. Tajima’s D test revealed no expanding or decreasing populations (0.15 < p < 0.89; Table 2). When the three western populations of the Alps are divided into a southern and northern component, Ne is higher, and H and π are slightly higher in the south than in the north.
Eighty-seven per cent of the total European haplotype variation is within population variation, whereas 13% is due to variation among the populations (Table 3). In the Alps, haplotype pairwise Φ PT values and pairwise nucleotide differences distinguish population D from the other three (Online Resource Table S4). According to the haplotype pairwise Φ PT values, populations of the Alps differ from most other European populations, but populations C and D could not be distinguished from two or three of the four northernmost Scandinavian populations. Populations N and NE, north and northeast of the Alps, are not distinguishable from GUD or EEU, and GUD, JMT, NNO, or EEU, respectively. Compared with the haplotype data, the pairwise nucleotide differences suggest somewhat less differentiation among the populations (Online Resource Table S4). The first three axes in PCoA analyses explain 77% of the variation among populations based on haplotype data and 61% for the nucleotide data. Population D differs from the other Alps populations (Fig. 4). According to the haplotype data, populations from the Alps appear close to each other and to N (especially for D), VAS (B), and several of the northernmost European populations. Nucleotide data suggest an affinity between D and several of the northern Scandinavian populations. With the alternative division of the Alps material, the northern and southern populations differ both from each other and from population N according to haplotype data (AMOVA: 0.035 > p > 0.004 for the three comparisons), but not according to nucleotide data.
The first three coordinates resulting from a Principal Coordinate Analysis (PCoA) based on the pairwise Φ PT values (a) or average numbers of pairwise nucleotide differences (b) among the eighteen European regional populations. In a, the first axis explains 35.6% of the variation in the data, the second 23.6%, and the third 17.4%, and the corresponding figures for b are 24.3, 19.5, and 17.3%. The shading of the dots corresponds with the indicated ranges along Coordinate axis 3. Acronyms for the different regions are explained in Table 1 and Fig. 1
A Mantel test based on inter-region Φ PT of all European populations suggested smaller inter-regional differences among more distant populations than among closely situated ones (Fig. 5) No such effect was found for the number of nucleotide differences (p = 0.203).
Thirty-four haplotypes occur in Scandinavia and fifteen haplotypes were private to this area, compared with 23 and 9, respectively, in the Alps. Resampling 73 specimens of the Scandinavian population 50 times yielded an average of 21.28 (95% confidence interval: 20.64–21.92) for the total number of haplotypes and 7.76 (7.25–8.27) private haplotypes for the entire Scandinavia.
Discussion
Within the Alps, the easternmost regional Rhytidium population differs statistically significantly from the other three populations. The two eastern populations are similar to populations in northern Scandinavia, suggesting a joint origin with the latter. The two western Alps populations appear isolated from the other studied European populations, and may have immigrated to the Alps from the S, SW, or W. Differences in intraspecific diversity between the Alps and Scandinavia are small; the slightly lower overall diversity in Scandinavia could result from the presence of isolated or marginal populations with a low diversity (Table 2).
The regional populations A–C in the Alps are similar to each other and differ from population D based on both haplotype and nucleotide data. The differentiation between A–C and D coincides approximately with the eastern major break zone for flowering plants around the Adige valley (Thiel-Egenter et al. 2011). Thus, the pattern found for Rhytidium in the east is consistent with the hypothesis that break zones of spore-dispersed organisms should mirror those of genotype patterns in flowering plants. Sampling of populations further to the W and SW is required to evaluate whether molecular variation in Rhytidium also reflects the Aoste valley break zone. Minor differences among populations A-C in the nucleotide data (Fig. 4b) and in haplotype composition (Fig. 2a) require a denser sampling for further evaluation.
Rhytidium in the Alps and their surroundings does currently not produce sporophytes, and explanations involving analogies with pollen dispersal in flowering plants may, therefore, seem implausible. This lack of Rhytidium sporophytes in central Europe is likely a new feature. Herbarium specimens as well as literature reports indicate several finds of sporophytes in a zone from SW Germany to E Austria during the 19th and early 20th centuries (Hedenäs 2015: Supporting information Fig. S1). Sex organs are still produced in the Alps (Hedenäs and Bisang, unpublished data), and most likely the present lack of sporophyte production is instead due to the absence of fertilization possibilities in this dioicous species. During re-colonization after the LGM and until traditional agriculture practices were abandoned, suitable habitat for Rhytidium was more common than presently in many areas of central Europe (cf., Meinunger and Schröder 2007). With more available habitat, male and female plants could more often grow close enough to each other for fertilization, i.e. within a few decimetres of each other (Bisang et al. 2004).
Dispersal limitations are probably weak in Rhytidium (see above, and Hedenäs 2015), and other factors must therefore explain the structuring of this species’ intraspecific variation. The geographical pattern within Rhytidium that we observe today could to a great extent have established early during the post-glacial colonization process (Hedenäs 2015), after which it was conserved according to the ‘founder takes all’ density-dependent principle (Waters et al. 2013). The relatively sharp differentiation between populations A–C and D in the Alps, seen both in the distribution of several individual haplotypes and for representatives of several haplotype lineages or groups (Fig. 2), is also expected by scenario ‘C’ of De Meester (2011). Under this scenario, small and efficiently dispersed species fill their niches in a landscape fast once they establish. Geographical barriers, such as the Adige valley break zone, cause the haplotype compositions on the two sides of the barrier to differ relatively strongly from each other.
The signals of haplotype and nucleotide data of the northern and southern partitions of the combined Alps regional Rhytidium populations A–C are conflicting, and the present results neither support nor refute post-glacial colonization separately from northern and southern marginal refugia (Schönswetter et al. 2005). The found haplotype differentiation between the N and S sides of the Alps possibly reflects a harsh early post-glacial climate of the inner portions of the mountains that served as a barrier to north–south dispersal. Over a period of c. 10,000 years ice remained and glaciers repeatedly partly melted and re-advanced (Ivy-Ochs et al. 2006, 2008). Once the climate of the inner mountains became more suitable, Rhytidium could more easily establish and disperse from one side to the other. However, then a high proportion of the suitable habitats were likely already occupied and diaspores would have difficulties in establishing, according to scenario ‘C’ of De Meester (2011) and in analogy with the pattern seen in Scandinavia (Hedenäs 2015). On the other hand, the lack of WSW-ENE differentiation could be an effect of repeated Rhytidium expansion and contraction in forelands that became ice-free c. 21,000 years BP, and thereafter passed through a number of climatic oscillations (Ivy-Ochs et al. 2006, 2008). During such fluctuations of variable intensities, smaller or larger areas of habitat suitable for Rhytidium must repeatedly have been lost and newly created. The repeated colonizations of newly created habitat could have counteracted the ‘founder takes all’ density-dependent principle (Waters et al. 2013) and have allowed a relatively mixed haplotype composition to develop in large areas.
Haplotype as well as nucleotide data indicate similarities between population D and the northernmost Scandinavian populations FNM, TRS, and NNO, but also RAG (in mid-Scandinavia; only nucleotide data), and between population C and the mid- to northern Scandinavian NNO, SNO, and GUD (nucleotide data). This is reflected by a substantial occurrence of members of lineages/groups ‘c’, ‘f’, and ‘h’ both in the Alps and in the northern portions of the Scandinavian mountain range or more generally in the north (Fig. 3). It also agrees with the Mantel test results from the haplotype data, which indicate that some of the geographically most distant populations are also most similar to each other, contrasting with larger differences between many populations that are located closer to each other (Fig. 5). Similarities between populations of the Alps or eastern Alps and those of the Scandinavian mountain range are known among flowering plants, such as Minuartia biflora (Schönswetter et al. 2006), Vaccinium uliginosum (Alsos et al. 2005), and eastern Alps populations of Ranunculus glacialis (Schönswetter et al. 2003). These studies sampled the entire Scandinavian mountain range relatively sparsely and did not find variation among different portions of the mountain range. However, for Rhytidium the similarities are primarily between northern Scandinavia and the eastern Alps, with mostly deviating populations in between. The southern portion of the Scandinavian peninsula was colonized by Rhytidium from the south, whereas the north was colonized from the northeast, following the retreating ice margins (Hedenäs 2015).
A direct north–south connection between the eastern Alps and northern Scandinavia is unlikely. The regional populations in between, most strikingly several of those in the lowlands, differ from those of the Alps as well as from those of the Scandinavian mountain range, especially in their haplotype compositions (Online Resource Table S4). This could be a result of survival of ancient local genotypes until the present in some lowland areas that were never covered by ice during the LGM, immigration from different source populations, and bottlenecks or founder effects, including ‘founder takes all’ consequences (Frankham et al. 2002; Waters et al. 2013). For example, the OEL population is known to have passed through periods with much less available habitat around 6 and 1.5 kA BP (Hedenäs 2015; Königsson 1968). Regional distinctiveness could also be due to restriction to clonal reproduction (Pfeiffer et al. 2006) and genetic drift due to low effective population sizes as was suggested for some Scandinavian Rhytidium populations (Hedenäs 2015).
Because the southern Scandinavian populations immigrated from south of the LGM ice shield, their haplotype compositions, which differ from those in northern Scandinavia, most likely reflect that of the LGM population of the western European lowlands (Hedenäs 2015). If the latter areas were occupied by populations of another composition than those of the far north, the similar ones found in the far north and the eastern Alps must have another origin. Most likely the latter two areas are connected through glacial refugia in the northeast or east, but to resolve this issue in more detail a much denser sampling than could be achieved so far is required for this enormous area (EEU; Table 1). A complicating factor is that numerous representatives of this putative source population have probably disappeared since the LGM in view of the current lack of Rhytidium in vast areas of Eastern Europe. Considering their distinctness from population D and lowland populations N and NE, the western Rhytidium populations of the Alps may have survived the LGM in refugia further to the west or southwest, but also to clarify this a more extensive sampling of the assumed refugial areas is required.
Conclusions
The three studied western regional populations of Rhytidium in the Alps are similar to each other and differ significantly from that in the ENE. Geographically, this differentiation coincides with the Adige valley break zone found for vascular plants. Rhytidium populations in Europe possibly originated in three major LGM refugia. (1) In E-NE Europe, which would explain similarities between the eastern Alps and northernmost Scandinavia. (2) In between the Scandinavian and Alps LGM ice shields, which would explain why southern mountain and lowland populations of Scandinavia and populations north of the Alps differ from those further north and south. (3) To the S, SW, and W of the Swiss Alps, which would explain why the studied western Alps populations differ from those in the east and display limited similarities to other populations. Because plants producing spores disperse much more efficiently than flowering plants, thus causing less distinct phylogeographic patterns, and since large areas in NE and SW Europe were insufficiently sampled to fully address the suggested origins (1) and (3), further investigations are still required to fully understand the phylogeography of Rhytidium in Europe.
The regional differentiation and the occurrence of numerous private Rhytidium haplotypes in the Alps as well as in other European regions show that conservation efforts, for bryophytes as well as for other organisms, must consider intraspecific variation if the aim is to conserve the entire biodiversity and not just rare species (Hedenäs 2016).
References
Alsos IG, Engelskjön T, Gielly L, Taberlet P, Brochmann C (2005) Impact of ice ages on circumpolar molecular diversity: insights from an ecological key species. Molec Ecol 14:2739–2753. doi:10.1111/j.1365-294X.2005.02621.x
Alsos IG, Alm T, Normand S, Brochmann C (2009) Past and future range shifts and loss of diversity in dwarf willow (Salix herbacea L.) inferred from genetics, fossils and modelling. Global Ecol Biogeogr 18:223–239
Bisang I, Ehrlén J, Hedenäs L (2004) Mate limited reproductive success in two dioicous mosses. Oikos 104:291–298. doi:10.1111/j.0030-1299.2004.12853.x
Brochmann C, Gabrielsen TM, Nordal I, Landvik JY, Elven R (2003) Glacial survival or tabula rasa? The history of North Atlantic biota revisited. Taxon 52:417–450
Bruen TC, Hervé P, Bryant D (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681. doi:10.1534/genetics.105.048975
Buckley HL, Paterson AM, Cruickshank RH et al (2013) The founder space race: a response to Waters et al. Trends Ecol Evol 28:189–190. doi:10.1016/j.tree.2013.01.005
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Molec Ecol 9:1657–1659. doi:10.1046/j.1365-294x.2000.01020.x
De Meester L (2011) A metacommunity perspective on the phylo- and biogeography of small organisms. In: Fontaneto D (ed) Biogeography of microscopic organisms. Cambridge University Press, Cambridge, pp 324–334
Dullinger F et al (2012) Post-glacial migration lag restricts range filling of plants in the European Alps. Global Ecol Biogeogr 21:829–840. doi:10.1111/j.1466-8238.2011.00732.x
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. doi:10.1111/j.1755-0998.2010.02847.x
Feliner GN, Rosselló JA (2007) Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Molec Phylog Evol 44:911–919. doi:10.1016/j.ympev.2007.01.013
Flø D, Hågvar S (2013) Aerial dispersal of invertebrates and mosses close to a receding alpine glacier in southern Norway. Arct Antarct Alp Res 45:481–490. doi:10.1657/1938-4246-45.4.481
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Fu Y (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Gugerli F, Holderegger R (2001) Nunatak survival, tabula rasa and the influence of the Pleistocene ice-ages on plant evolution in mountain areas. Trends Plant Sci 6:397–398. doi:10.1016/S1360-1385(01)02053-2
Guo L, Ma F, Wei F, Fanella B, Allen DK, Wang X (2014) Cytosolic phosphorylating glyceraldehyde-3-phosphate dehydrogenase affect Arabidopsis cellular metabolism and promote seed oil accumulation. Plant Cell 26:3023–3035. doi:10.1105/tpc.114.126946
Hedenäs L (2010) Phylogeography and origin of European Sanionia uncinata (Amblystegiaceae, Bryophyta). System Biodivers 8:177–191. doi:10.1080/14772001003767600
Hedenäs L (2012) Molecular differentiation within European Cratoneuron filicinum, and differences towards Asiatic and American populations. Plant Syst Evol 298:937–945. doi:10.1007/s00606-012-0603-y
Hedenäs L (2014) Southern Scandinavian lowland populations of Rhytidium rugosum (Bryophyta, Rhytidiaceae) differ significantly from those in the mountains. J Bryol 36:1–14. doi:10.1179/1743282013Y.0000000080
Hedenäs L (2015) Rhytidium rugosum (Bryophyta) colonized Scandinavia from at least two glacial refugial source populations. Bot J Linn Soc 179:635–657. doi:10.1111/boj.12341
Hedenäs L (2016) Intraspecific diversity matters in bryophyte conservation–internal transcribed spacer and rpl16 G2 intron variation in European mosses. J Bryol 38:173–182. doi:10.1080/03736687.2016.1145522
Hedenäs L, Eldenäs P (2007) Cryptic speciation, habitat differentiation, and geography in Hamatocaulis vernicosus (Calliergonaceae, Bryophyta). Plant Syst Evol 268:131–145. doi:10.1007/s00606-007-0529-y
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913. doi:10.1038/35016000
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi:10.1093/molbev/msj030
Ivy-Ochs S et al (2006) The timing of glacier advances in the northern European Alps based on surface exposure dating with cosmogenic 10Be, 26Al, 36Cl, and 21Ne. Geol Soc Am Spec Pap 415:43–60. doi:10.1130/2006.2415(04)
Ivy-Ochs S et al (2008) Chronology of the last glacial cycle in the European Alps. J Quaternary Sci 23:559–573. doi:10.1002/jqs.1202
Jaarola M, Tegelström H, Fredga K (1999) Colonization history in Fennoscandian rodents. Biol J Linn Soc 68:113–127. doi:10.1111/j.1095-8312.1999.tb01161.x
Koch MA, Kiefer C, Ehrich D, Vogel J, Brochmann C, Mummenhoff K (2006) Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae). Molec Ecol 15:825–839. doi:10.1111/j.1365-294X.2005.02848.x
Königsson L-K (1968) The Holocene history of the Great Alvar of Öland. Acta Phytogeogr Suec 55:1–172
Kophimai Y (2013) Population genetics of the fen specialist moss Scorpidium cossonii in northeastern Switzerland, PhD thesis, Universität Bern
Košnar J, Herbstová M, Kolář F, Koutecký P, Kučera J (2012) A case of intragenomic ITS variation in bryophytes: assessment of gene flow and role of plyploidy in the origin of European taxa of the Tortula muralis (Musci: Pottiaceae) complex. Taxon 61:709–720
Kyrkjeeide MO, Stenøien HK, Flatberg KI, Hassel K (2014) Glacial refugia and post-glacial colonization patterns in European bryophytes. Lindbergia 37:47–59
Lambeck K, Purcell A, Zhao J, Svensson N-O (2010) The Scandinavian ice sheet: from MIS 4 to the end of the Last Glacial Maximum. Boreas 39:410–435. doi:10.1111/j.1502-3885.2010.00140.x
Meinunger L, Schröder W (2007) Verbreitungsatlas der Moose Deutschlands, vol 3. Regensburgische Botanische Gesellschaft, Regensburg
Mikulášková E, Hájek M, Veleba A, Johnson MG, Hájek T, Shaw JA (2014) Local adaptations in bryophytes revisited: the genetic structure of the calci-tolerant peatmoss Sphagnum warnstorfii along geographic and pH gradients. Ecol Evol 5:229–242. doi:10.1002/ece3.1351
Miller NG, Ambrose LJH (1976) Growth in culture of wind-blown bryophyte gametophyte fragments from Arctic Canada. Bryologist 79:55–63. doi:10.2307/3241866
Ochyra R, Szmajda P (1983) M 628. Rhytidium rugosum (Hedw.) Kindb. In: Szweykowski J, Wojterski T (eds) Atlas of the geographical distribution of spore plants in Poland. Series V. Mosses (Musci). Part I. Panstwowe Wydawnictwo Naukowe, Warzawa, pp 29–31
Parducci L et al (2012) Glacial survival of boreal trees in northern Scandinavia. Science 335:1083–1086. doi:10.1126/science.1216043
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teching and research. Mol Ecol Notes 6:288–295. doi:10.1111/j.1471-8286.2005.01155.x
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pfeiffer T, Fritz S, Stech M, Frey W (2006) Vegetative reproduction and clonal diversity in Rhytidium rugosum (Rhytidiaceae, Bryopsida) inferred by morpho-anatomical and molecular analyses. J Plant Res 119:125–135. doi:10.1007/s10265-005-0255-x
Ramírez-Soriano A, Ramos-Onsins SE, Rozas J, Calafell F, Navarro A (2008) Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics 179:555–567. doi:10.1534/genetics.107.083006
Schönswetter P, Paun O, Tribsch A, Niklfeld H (2003) Out of the Alps: colonization of northern Europe by east Alpine populations of the Glacier Buttercup Ranunculus glacialis L. (Ranunculaceae). Molec Ecol 12:3373–3381. doi:10.1046/j.1365-294X.2003.01984.x
Schönswetter P, Stehlik I, Holderegger R, Tribsch A (2005) Molecular evidence for glacial refugia of mountain plants in the European Alps. Molec Ecol 2005:3547–3555. doi:10.1111/j.1365-294X.2005.02683.x
Schönswetter P, Popp M, Brochmann C (2006) Rare arctic-alpine plants of the European Alps have different immigration histories: the snow bed species Minuartia biflora and Ranunculus pygmaeus. Molec Ecol 15:709–720. doi:10.1111/j.1365-294X.2006.02821.x
Shaw AJ, McDaniel SF, Werner O, Ros RM (2002) New frontiers in bryology and lichenology. Phylogeogr Phylodemogr Bryol 105:373–383. doi:10.1639/0007-2745(2002)105[0373:PAP]2.0.CO;2
Sirover MA (1999) New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta (BBA) Protein Struct Mol Enzymol 1432:159–184. doi:10.1016/S0167-4838(99)00119-3
Stech M (1999) Molekulare Systematik haplolepider Laubmoose (Dicranaceae, Bryopsida). PhD thesis, Freie Universität Berlin, Berlin
Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson J-F (1998) Comparative phylogeography and postglacial colonization routes in Europe. Molec Ecol 7:453–464. doi:10.1046/j.1365-294x.1998.00289.x
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Thiel-Egenter C et al (2011) Break zones in the distributions of alleles and species in alpine plants. J Biogeogr 38:772–782. doi:10.1111/j.1365-2699.2010.02441.x
Vorren TO, Vorren K-D, Aasheim O, Dahlgren KIT, Forwick M, Hassel K (2013) Palaeoenvironment in northern Norway between 22.2 and 14.5 cal. ka BP. Boreas 42:876–895. doi:10.1111/bor.12013
Walser J-C, Holderegger R, Gugerli F, Hoebee SE, Scheidegger C (2005) Microsatellites reveal regional population differentiation and isolation in Lobaria pulmonaria, an epiphytic lichen. Molec Ecol 14:457–467. doi:10.1111/j.1365-294X.2004.02423.x
Waters JM, Fraser CI, Hewitt GM (2013) Founder takes all: density-dependent processes structure biodiversity. Trends Ecol Evol 28:78–85. doi:10.1016/j.tree.2012.08.024
Westergaard KB, Alsos IG, Popp M, Engelskjön T, Flatberg KI, Brochmann C (2011) Glacial survival may matter after all: nunatak signatures in the rare European populations of two west-arctic species. Molec Ecol 20:376–393. doi:10.1111/j.1365-294X.2010.04928.x
Widmer I, dal Grande F, Excoffier L, Holderegger R, Keller C, Mikryukov VS, Scheidegger C (2012) European phylogeography of the epiphytic lichen fungus Lobaria pulmonaria and its green algal symbiont. Molec Ecol 21:5827–5844. doi:10.1111/mec.12051
Wilkinson DM, Koumoutsaris S, Mitchell EAD, Bey I (2012) Modelling the effect of size on the aerial dispersal of microorganisms. J Biogeogr 39:89–97. doi:10.1111/j.1365-2699.2011.02569.x
Zoller S, Lutzoni F, Scheidegger C (1999) Genetic variation within and among populations of the threatened lichen Lobaria pulmonaria in Switzerland and implications for its conservation. Molec Ecol 8:2049–2059. doi:10.1046/j.1365-294x.1999.00820.x
Acknowledgements
I thank Rasa Bukontaite for efficient laboratory work, and the curators of BOZ, G, GZU, LI, Z, and W, as well as H. Hofmann, M. Meier, L. Miserere, F. Roloff, and N. Schnyder for loans of Rhytidium. Comments by two reviewers significantly improved this paper. ‘Magnus Bergvalls stiftelse’ (No. 2015-00657) funded this study.
Author information
Authors and Affiliations
Contributions
Lars Hedenäs designed the study, executed all portions except the molecular laboratory work, analysed the data, wrote the text and produced the illustrations.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hedenäs, L. Phylogeography of Alpine populations of Rhytidium rugosum (Bryophyta) in a European context. Alp Botany 127, 197–209 (2017). https://doi.org/10.1007/s00035-017-0191-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00035-017-0191-0