Abstract
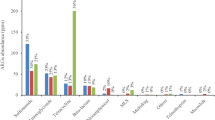
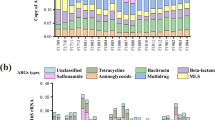
Antibiotic resistance genes (ARGs) are emerging contaminants that pose a potential threat to human health worldwide. Urban wastewater treatment plants (WWTPs) are a main source of both antibiotic-resistant bacteria and ARGs released into the environment. Nevertheless, the propagation of ARGs and their underlying mechanisms and the dynamics of mobile genetic elements (MGEs) in WWTPs have rarely been investigated in South Korea. In this study, shotgun metagenomic analysis was used to identify comprehensive ARGs and their mechanisms, bacterial communities, and MGEs from 4 configurations with 2 activated sludge (AS) and 2 anaerobic digestion sludge (ADS) samples. A total of 181 ARG subtypes belonging to 22 ARG types were broadly detected, and the ARG abundances in the AS samples were 1.3–2.0 orders of magnitude higher than in the ADS samples. Multidrug and bacitracin resistance genes were the predominant ARG types in AS samples, followed by ARGs against sulfonamide, tetracycline, and β-lactam. However, the composition of ARG types in ADS samples was significantly changed. The abundance of multidrug and β-lactam resistance genes was drastically reduced in the ADS samples. The resistance genes of MLS were the predominant, followed by ARGs against sulfonamide and tetracycline in the ADS samples. In addition, plasmids were the dominant MGEs in the AS samples, while integrons (intI1) were the dominant MGEs in the ADS samples. These results provide valuable information regarding the prevalence of ARG types and MGEs and the difference patterns between the AS and ADS systems.
Similar content being viewed by others
References
Allen, H.K., Donato, J., Wang, H.H., Cloud-Hansen, K.A., Davies, J., and Handelsman, J. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol.8, 251.
Ariesyady, H.D., Ito, T., and Okabe, S. 2007. Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res.41, 1554–1568.
Baquero, F., Martínez, J.L., and Cantón, R. 2008. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol.19, 260–265.
Bouki, C., Venieri, D., and Diamadopoulos, E. 2013. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf.91, 1–9.
Chen, Q., An, X., Li, H., Su, J., Ma, Y., and Zhu, Y.G. 2016. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int.92–93, 1–10.
Chen, B., Yang, Y., Liang, X., Yu, K., Zhang, T., and Li, X. 2013. Metagenomic profiles of antibiotic resistance genes (ARGs) between human impacted estuary and deep ocean sediments. Environ. Sci. Technol.47, 12753–12760.
Christgen, B., Yang, Y., Ahammad, S.Z., Li, B., Rodriquez, D.C., Zhang, T., and Graham, D.W. 2015. Metagenomics shows that low-energy anaerobic-aerobic treatment reactors reduce antibiotic resistance gene levels from domestic wastewater. Environ. Sci. Technol.49, 2577–2584.
Devarajan, N., Laffite, A., Graham, N.D., Meijer, M., Prabakar, K., Mubedi, J.I., Elongo, V., Mpiana, P.T., Ibelings, B.W., Wildi, W., et al. 2015. Accumulation of clinically relevant antibiotic-resistance genes, bacterial load, and metals in freshwater lake sediments in central Europe. Environ. Sci. Technol.49, 6528–6537.
Fang, H., Huang, K., Yu, J., Ding, C., Wang, Z., Zhao, C., Yuan, H., Wang, Z., Wang, S., Hu, J., et al. 2019. Metagenomic analysis of bacterial communities and antibiotic resistance genes in the Eriocheir sinensis freshwater aquaculture environment. Chemosphere224, 202–211.
Gao, P., Munir, M., and Xagoraraki, I. 2012. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal waste-water treatment plant. Sci. Total Environ.421–422, 173–183.
Garcia-Peña, E.I., Parameswaran, P., Kang, D.W., Canul-Chan, M., and Krajmalnik-Brown, R. 2011. Anaerobic digestion and co-digestion processes of vegetable and fruit residues: Process and microbial ecology. Bioresour. Technol.102, 9447–9455.
Gaze, W.H., Zhang, L., Abdouslam, N.A., Hawkey, P.M., Calvo-Bado, L., Royle, J., Brown, H., Davis, S., Kay, P., Boxall, A.B.A., et al. 2011. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J.5, 1253.
Ghosh, S., Ramsden, S.J., and LaPara, T.M. 2009. The role of anaerobic digestion in controlling the release of tetracycline resistance genes and class 1 integrons from municipal wastewater treatment plants. Appl. Microbiol. Biotechnol.84, 791–796.
Gillings, M.R., Gaze, W.H., Pruden, A., Smalla, K., Tiedje, J.M., and Zhu, Y.G. 2014. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J.9, 1269.
Guo, J., Li, J., Chen, H., Bond, P.L., and Yuan, Z. 2017. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res.123, 468–478.
Gupta, S.K., Shin, H., Han, D., Hur, H.G., and Unno, T. 2018. Metagenomic analysis reveals the prevalence and persistence of antibiotic-and heavy metal-resistance genes in wastewater treatment plant. J. Microbiol.56, 408–415.
Hsu, J.T., Chen, C.Y., Young, C.W., Chao, W.L., Li, M.H., Liu, Y.H., Lin, C.M., and Ying, C. 2014. Prevalence of sulfonamide-resistant bacteria, resistance genes and integron-associated horizontal gene transfer in natural water bodies and soils adjacent to a swine feedlot in northern Taiwan. J. Hazard. Mater.277, 34–43.
Jia, S., Shi, P., Hu, Q., Li, B., Zhang, T., and Zhang, X.X. 2015. Bacterial community shift drives antibiotic resistance promotion during drinking water chlorination. Environ. Sci. Technol.49, 12271–12279.
Ju, F., Li, B., Ma, L., Wang, Y., Huang, D., and Zhang, T. 2016. Antibiotic resistance genes and human bacterial pathogens: Co-occurrence, removal, and enrichment in municipal sewage sludge digesters. Water Res.91, 1–10.
Kim, S. and Aga, D.S. 2007. Potential ecological and human health impacts of antibiotics and antibiotic-resistant bacteria from waste-water treatment plants. J. Toxicol. Environ. Heal. Part B10, 559–573.
Kolz, A.C., Ong, S.K., and Moorman, T.B. 2005. Sorption of tylosin onto swine manure. Chemosphere60, 284–289.
Kristiansson, E., Fick, J., Janzon, A., Grabic, R., Rutgersson, C., Weijdegård, B., Söderström, H., and Larsson, D.G.J. 2011. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One6, e17038.
LaPara, T.M., Burch, T.R., McNamara, P.J., Tan, D.T., Yan, M., and Eichmiller, J.J. 2011. Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor. Environ. Sci. Technol.45, 9543–9549.
Li, B., Yang, Y., Ma, L., Ju, F., Guo, F., Tiedje, J.M., and Zhang, T. 2015. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J.9, 2490.
Lu, X., Zhang, X.X., Wang, Z., Huang, K., Wang, Y., Liang, W., Tan, Y., Liu, B., and Tang, J. 2015. Bacterial pathogens and community composition in advanced sewage treatment systems revealed by metagenomics analysis based on high-throughput sequencing. PLoS One10, e0125549.
Ma, Y., Wilson, C.A., Novak, J.T., Riffat, R., Aynur, S., Murthy, S., and Pruden, A. 2011. Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class I integrons. Environ. Sci. Technol.45, 7855–7861.
Manaia, C.M., Rocha, J., Scaccia, N., Marano, R., Radu, E., Biancullo, F., Cerqueira, F., Fortunato, G., Iakovides, I.C., Zammit, I., et al. 2018. Antibiotic resistance in wastewater treatment plants: Tackling the black box. Environ. Int.115, 312–324.
Mao, D., Yu, S., Rysz, M., Luo, Y., Yang, F., Li, F., Hou, J., Mu, Q., and Alvarez, P.J.J. 2015. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res.85, 458–466.
McCarty, P.L., Bae, J., and Kim, J. 2011. Domestic wastewater treatment as a net energy producer-can this be achieved? Environ. Sci. Technol.45, 7100–7106.
Munir, M., Wong, K., and Xagoraraki, I. 2011. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res.45, 681–693.
Pál, C., Papp, B., and Lercher, M. J. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet.37, 1372–1375.
Park, J., Han, E., Lee, S.O., and Kim, D.S. 2017. Antibiotic use in South Korea from 2007 to 2014: A health insurance database-generated time series analysis. PLoS One12, e0177435.
Pruden, A., Larsson, D.J., Amézquita, A., Collignon, P., Brandt, K.K., Graham, D.W., Lazorchak, J.M., Suzuki, S., Silley, P., Snape, J.R., et al. 2013. Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ. Heal. Persp.121, 878–885.
Rizzo, L., Manaia, C., Merlin, C., Schwartz, T., Dagot, C., Ploy, M.C., Michael, I., and Fatta-Kassinos, D. 2013. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ.447, 345–360.
Su, J.Q., Wei, B., Ou-Yang, W.Y., Huang, F.Y., Zhao, Y., Xu, H.J., and Zhu, Y.G. 2015. Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol.49, 7356–7363.
Traversi, D., Villa, S., Lorenzi, E., Degan, R., and Gilli, G. 2012. Application of a real-time qPCR method to measure the methanogen concentration during anaerobic digestion as an indicator of biogas production capacity. J. Environ. Manage.111, 173–177.
Truong, D.T., Franzosa, E.A., Tickle, T.L., Scholz, M., Weingart, G., Pasolli, E., Tett, A., Huttenhower, C., and Segata, N. 2015. Meta-PhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods12, 902–903.
Wagner, M. and Loy, A. 2002. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol.13, 218–227.
Wang, Z., Zhang, X.X., Huang, K., Miao, Y., Shi, P., Liu, B., Long, C., and Li, A. 2013. Metagenomic profiling of antibiotic resistance genes and mobile genetic elements in a tannery wastewater treatment plant. PLoS One8, e76079.
Wu, Y., Cui, E., Zuo, Y., Cheng, W., Rensing, C., and Chen, H. 2016. Influence of two-phase anaerobic digestion on fate of selected antibiotic resistance genes and class I integrons in municipal waste-water sludge. Bioresour. Technol.211, 414–421.
Xu, J., Xu, Y., Wang, H., Guo, C., Qiu, H., He, Y., Zhang, Y., Li, X., and Meng, W. 2015. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere119, 1379–1385.
Yang, Y., Jiang, X., Chai, B., Ma, L., Li, B., Zhang, A., Cole, J.R., Tiedje, J.M., and Zhang, T. 2016. ARGs-OAP: online analysis pipeline for antibiotic resistance genes detection from metagenomic data using an integrated structured ARG-database. Bioinformatics32, 2346–2351.
Yang, Y., Li, B., Ju, F., and Zhang, T. 2013. Exploring variation of antibiotic resistance genes in activated sludge over a four-year period through a metagenomic approach. Environ. Sci. Technol.47, 10197–10205.
Yang, Y., Li, B., Zou, S., Fang, H.H.P., and Zhang, T. 2014. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res.62, 97–106.
Yin, X.L., Jiang, X.T., Chai, B.L., Li, L.G., Yang, Y., Cole, J.R., Tiedje, J.M., and Zhang, T. 2018. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics34, 2263–2270.
Yoo, K., Yoo, H., Lee, J.M., Shukla, S.K. and Park, J. 2018. Classification and regression tree approach for prediction of potential hazards of urban airborne bacteria during Asian dust events. Sci. Rep.8, 11823.
Zhang, J., Chen, M., Sui, Q., Tong, J., Jiang, C., Lu, X., Zhang, Y., and Wei, Y. 2016a. Impacts of addition of natural zeolite or a nitrification inhibitor on antibiotic resistance genes during sludge composting. Water Res.91, 339–349.
Zhang, J., Chen, M., Sui, Q., Wang, R., Tong, J., and Wei, Y. 2016b. Fate of antibiotic resistance genes and its drivers during anaerobic co-digestion of food waste and sewage sludge based on microwave pretreatment. Bioresour. Technol.217, 28–36.
Zhang, T., Yang, Y., and Pruden, A. 2015. Effect of temperature on removal of antibiotic resistance genes by anaerobic digestion of activated sludge revealed by metagenomic approach. Appl. Microbiol. Biotechnol.99, 7771–7779.
Zhang, T., Zhang, X.X., and Ye, L. 2011. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS One6, e26041.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2018R1-A6A1A08025348).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Yoo, K., Yoo, H., Lee, J. et al. Exploring the antibiotic resistome in activated sludge and anaerobic digestion sludge in an urban wastewater treatment plant via metagenomic analysis. J Microbiol. 58, 123–130 (2020). https://doi.org/10.1007/s12275-020-9309-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-020-9309-y