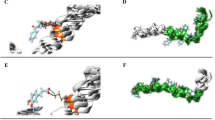
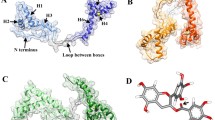
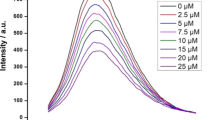
Abstract
To inhibit hIAPP aggregation and reduce toxicity of its oligomers are one of the potential strategies for the treatment of Type 2 diabetes (T2D). It has been reported that there is an effective inhibitory effect on hIAPP aggregation by five natural flavonoids, including Genistein, Rutin, Quercetin, Epigallocatechin gallate (EGCG), and Silibinin, which are widely found in our daily food. However, the detailed mechanisms to inhibit hIAPP aggregation remain unclear. Here, we explore the mechanisms of the five flavonoids against hIAPP aggregation by molecular docking and molecular dynamics simulations. We show that these flavonoids can disaggregate Chain A and Chain B of hIAPP to reduce the extended conformation by binding with two regions of hIAPP, Leu12–Ala13–Asn14 and Asn31–Val32–Gly33–Ser34–Asn35, with the inhibitory ability of Genistein > Rutin > Quercetin > EGCG > Silibinin. These five compounds exhibit a common mechanism for disaggregation of the hIAPP pentamer; that is, they loosen the two nearest peptide chains to potentially destroy the hIAPP oligomer. Mutations of eight key residues remarkably affected by the flavonoids indicate that the secondary structures of the hIAPP pentamer change from β-sheet to be random coil, thereby to destroy its structural stability; moreover, the 28th (Ser), 12th (Leu) and 32nd (Val) amino acids exhibit significant effects on structural stability of the hIAPP pentamer, providing an important hint that these amino acids can be considered as potential targets for design of new candidate inhibitors against hIAPP oligomers. This work is beneficial to understanding of mechanism of these inhibits against hIAPP aggregation and will facilitate screening, modification, and design of new inhibitors.






Similar content being viewed by others
References
Hoskin MA, Bray GA, Hattaway K, Khare-Ranade PA, Pomeroy J, Semler LN, Weinzierl VA, Wylie-Rosett J (2014) Prevention of diabetes through the lifestyle intervention: lessons learned from the diabetes prevention program and outcomes study and its translation to practice. Curr Nutr Rep 3(4):364–378. https://doi.org/10.1007/s13668-014-0094-2
Oyebode OA, Erukainure OL, Chukwuma CI, Ibeji CU, Koorbanally NA, Islam S (2018) Boerhaavia diffusa inhibits key enzymes linked to type 2 diabetes in vitro and in silico; and modulates abdominal glucose absorption and muscle glucose uptake ex vivo. Biomed Pharmacother 106:1116–1125. https://doi.org/10.1016/j.biopha.2018.07.053
Saumya M, Subin EK, Suchithra TV (2019) Network analysis of MPO and other relevant proteins involved in diabetic foot Ulcer and other diabetic complications. Interdiscip Sci Comput Life Sci 11(2):180–190. https://doi.org/10.1007/s12539-017-0258-z
Cho NH, Shaw JE, Karuranga S, Huang Y, da Rocha Fernandes JD, Ohlrogge AW, Malanda B (2018) IDF Diabetes atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract 138:271–281. https://doi.org/10.1016/j.diabres.2018.02.023
Westermark P, Andersson A, Westermark GT (2011) Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol Rev 91(3):795–826. https://doi.org/10.1152/physrev.00042.2009
Mukherjee A, Morales-Scheihing D, Butler PC, Soto C (2015) Type 2 diabetes as a protein misfolding disease. Trends Mol Med 21(7):439–449. https://doi.org/10.1016/j.molmed.2015.04.005
Selkoe DJ (2003) Folding proteins in fatal ways. Nature 426(6968):900–904. https://doi.org/10.1038/nature02264
Lorenzo A, Razzaboni B, Weir GC, Yankner BA (1994) Pancreatic islet cell toxicity of amylin associated with type-2 diabetes mellitus. Nature 368(6473):756–760. https://doi.org/10.1038/368756a0
Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416(6880):507–511. https://doi.org/10.1038/416507a
Aitken JF, Loomes KM, Riba-Garcia I, Unwin RD, Prijic G, Phillips AS, Phillips ARJ, Wu D, Poppitt SD, Ding K, Barran PE, Dowsey AW, Cooper GJS (2017) Rutin suppresses human-amylin/hIAPP misfolding and oligomer formation in vitro, and ameliorates diabetes and its impacts in human-amylin/hIAPP transgenic mice. Biochem Biophys Res Commun 482(4):625–631. https://doi.org/10.1016/j.bbrc.2016.11.083
Cheng B, Gong H, Li X, Sun Y, Zhang X, Chen H, Liu X, Zheng L, Huang K (2012) Silibinin inhibits the toxic aggregation of human islet amyloid polypeptide. Biochem Biophys Res Commun 419(3):495–499. https://doi.org/10.1016/j.bbrc.2012.02.042
Ren B, Liu Y, Zhang Y, Cai Y, Gong X, Chang Y, Xu L, Zheng J (2018) Genistein: a dual inhibitor of both amyloid beta and human islet amylin peptides. ACS Chem Neurosci 9(5):1215–1224. https://doi.org/10.1021/acschemneuro.8b00039
López LC, Varea O, Navarro S, Carrodeguas JA, Sanchez de Groot N, Ventura S, Sancho J (2016) Benzbromarone, quercetin, and folic acid inhibit amylin aggregation. Int J Mol Sci 17(6):964
Xu ZX, Ma GL, Zhang Q, Chen CH, He YM, Xu LH, Zhou GR, Li ZH, Yang HJ, Zhou P (2017) Inhibitory mechanism of epigallocatechin gallate on fibrillation and aggregation of amidated human islet amyloid polypeptide. Chemphyschem 18(12):1611–1619. https://doi.org/10.1002/cphc.201700057
Simon L, Imane A, Srinivasan KK, Pathak L, Daoud I (2017) In silico drug-designing studies on flavanoids as anticolon Cancer Agents: pharmacophore mapping, molecular docking, and monte carlo method-based qsar modeling. Interdiscip Sci Comput Life Sci 9(3):445–458. https://doi.org/10.1007/s12539-016-0169-4
Moore S, Sonar K, Bharadwaj P, Deplazes E, Mancera R (2018) Characterisation of the structure and oligomerisation of islet amyloid polypeptides (IAPP): a review of molecular dynamics simulation studies. Molecules 23(9):2142
Luca S, Yau WM, Leapman R, Tycko R (2007) Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry 46(47):13505–13522. https://doi.org/10.1021/bi701427q
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2016) PubChem Substance and Compound databases. Nucleic Acids Res 44(D1):D1202–D11213. https://doi.org/10.1093/nar/gkv951
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2010) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Rosenman DJ, Wang C, Garcia AE (2016) Characterization of abeta monomers through the convergence of ensemble properties among simulations with multiple force fields. J Phys Chem B 120(2):259–277. https://doi.org/10.1021/acs.jpcb.5b09379
Liang G, Zhao J, Yu X, Zheng J (2013) Comparative molecular dynamics study of human islet amyloid polypeptide (IAPP) and rat IAPP oligomers. Biochemistry 52(6):1089–1100. https://doi.org/10.1021/bi301525e
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Huang J, MacKerell AD Jr (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem 34(25):2135–2145. https://doi.org/10.1002/jcc.23354
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26(16):1781–1802. https://doi.org/10.1002/jcc.20289
Andersena HC (1983) Rattle: a “velocity” version of the shake algorithm for molecular dynamics calculations. J Comput Phys 52(1):24–34
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23(3):327–341
Miyamoto S, Kollman PA (1992) Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 13(8):952–962
Laskowski RA, Swindells MB (2011) LigPlot + : multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51(10):2778–2786. https://doi.org/10.1021/ci200227u
Khan MKA, Akhtar S, Arif JM (2018) Structural Insight into the Mechanism of Dibenzo[a, l]pyrene and Benzo[a]pyrene-Mediated cell proliferation using molecular docking simulations. Interdiscip Sci Comput Life Sci 10(4):653–673. https://doi.org/10.1007/s12539-017-0226-7
Qiao Y, Zhang MZ, Liang YN, Zheng J, Liang GZ (2017) A computational study of self-assembled hexapeptide inhibitors against amyloid-beta (A beta) aggregation. Phys Chem Chem Phys 19(1):155–166. https://doi.org/10.1039/c6cp07341g
Wilcox G (2005) Insulin and insulin resistance. Clin Biochem Rev 26(2):19–39
Buchanan LE, Dunkelberger EB, Tran HQ, Pin-Nan C, Chi-Cheng C, Ping C, Raleigh DP, Pablo JJ, De Nowick JS, Zanni MT (2013) Mechanism of IAPP amyloid fibril formation involves an intermediate with a transient β-sheet. Proc Natl Acad Sci USA 110(48):19285–19290
Fan HM, Gu RX, Wang YJ, Pi YL, Zhang YH, Xu Q, Wei DQ (2015) Destabilization of Alzheimer’s Abeta42 protofibrils with a novel drug candidate wgx-50 by molecular dynamics simulations. J Phys Chem B 119(34):11196–11202. https://doi.org/10.1021/acs.jpcb.5b03116
Choudhary AK (2018) Aspartame: should Individuals with Type II Diabetes be Taking it? Curr Diabetes Rev 14(4):350–362. https://doi.org/10.2174/1573399813666170601093336
Fernández-Gómez I, Sablón-Carrazana M, Bencomo-Martínez A, Domínguez G, Lara-Martínez R, Altamirano-Bustamante NF, Jiménez-García LF, Pasten-Hidalgo K, Castillo-Rodríguez RA, Altamirano P (2018) Diabetes drug discovery: hIAPP1–37 polymorphic amyloid structures as novel therapeutic targets. Molecules 23(3):686
Li J, Fu A, Zhang L (2019) An overview of scoring functions used for protein-ligand interactions in molecular docking. Interdiscip Sci Comput Life Sci 11(2):320–328. https://doi.org/10.1007/s12539-019-00327-w
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31571782 and 31771975) and the Natural Science Foundation of Chongqing CSTC (No. cstc2018jcyjAX0765).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Y., Lv, Y., Jin, L. et al. Revealing the Mechanism of EGCG, Genistein, Rutin, Quercetin, and Silibinin Against hIAPP Aggregation via Computational Simulations. Interdiscip Sci Comput Life Sci 12, 59–68 (2020). https://doi.org/10.1007/s12539-019-00352-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-019-00352-9