Abstract
The aim of this study was to assess potential harmful lethal and sublethal effects of commercial plant protection product containing Bacillus amyloliquefaciens QST 713 (formerly subtilis) spores on winter adult honeybees according to OECD 245 (2017). Caged winter honeybees fed 10 days with a sucrose solution (50% w/w) ad libitum spiked with a maximum application field rate of 1/1 and its 1/10 showed no dose-dependent mortality under laboratory conditions; the No Observed Effect Concentration (NOEC) appears to be higher than or equal 4 × 107 colony-forming unit (CFU)/ml diet, resp. 4 × 1010 CFU/l diet. Although the results of Denaturing Gradient Gel Electrophoresis (DGGE) analysis showed no significant changes in the composition of gut bacterial community between untreated control and experimental groups after 10 days of chronic exposure, the expression of genes encoding antimicrobial peptides was decreased in one/both treated groups in comparison with untreated control group; significantly lower results of expression were detected in genes for Apidaecin, Defensin-1, and Hymenoptaecin. Despite no direct lethal effect of commercial plant protection product containing Bacillus amyloliquefaciens, QST 713 was observed in adult bees following chronic exposure; the decrease in some immunity parameters observed in tested winter bees may potentially impair bee colony health and survival.
Similar content being viewed by others
1 Introduction
The market for biopesticides has been reportedly growing at an average annual rate of 15% since 2010 (Frabotta 2014). Over the coming years, the growth is expected to be highest in regions of North and Latin America, Europe, and Asia (Olson 2015).
In the EU, the approval of microbial active substances in plant protection products (PPPs) is done at the strain/isolate level under the European Regulation (EC) No. 1107/2009. A microorganism is defined as any microbiological entity, including lower fungi and viruses, cellular or non-cellular, capable of replication or of transferring genetic material.
The commercial PPPs containing spores of Bacillus amyloliquefaciens QST 713 (formerly subtilis) as the active ingredient are classified as microbial fungicides. B. amyloliquefaciens strain QST 713 is a rod-shaped, Gram-positive, aerobic bacterium which was first isolated from soil from a peach tree orchard in Fresno County, California (USA), in 1995 and was first classified as Bacillus subtilis QST 713. Due to the close relationship and physiological similarity of the species, data on B. amyloliquefaciens can be used from data on B. subtilis (EFSA 2018). The bacteria possess significant efficacy against a broad spectrum of economically important fungal and bacterial diseases (incl. grey mould, powdery mildew early and late blight, fire blight, scab, sour rot, bacterial spot and walnut blight, etc.) in fruit and vegetable production (EPA 2006).
According to valid EU-agreed Good Agriculture Praxis directed use of Serenade® ASO, calculated concentration of B. amyloliquefaciens strain QST 713 in the spray solution for application in strawberries and grapevines is expected as the highest concentration agreed in EU (SANCO 2006; EFSA 2018). Field application in grapevines with BBCH (the abbreviation BBCH derives from Biologische Bundesanstalt, Bundessortenamt, and CHemical industry) up to 89 (berries ripe for harvest) is considered as a worst-case scenario for pollinators. Bee-attractive blossom weeds found in vineyards exposed during the application of Serenade® ASO gives us an assumption that the risk to non-target pollinators (honeybees included) cannot be excluded, as the application itself can potentially result in exposure of pollinators either through direct over-spray, or by contact with “spores” on plants while pollinators are foraging for food.
To that time, there is no scientific paper dealing with potential harmful lethal or sublethal effects of commercial product or technical product containing spores of B. amyloliquefaciens strain QST 713 as the active ingredient on non-target pollinators. Some assessment reports from e.g. EPA or EFSA can be freely found online (EPA 2006; EFSA 2018). According to EFSA’s Renewal assessment report, slight toxic effects of Bacillus amyloliquefaciens QST 713 (formerly subtilis) to honeybees from technical Bacillus amyloliquefaciens QST 713 or the product Serenade® ASO have been proven (EFSA 2018).
Most of the published studies are focused on Bacillus thuringiensis, quite commonly used active ingredient of commercial PPPs used in biological agriculture as insecticide. In vitro susceptibility of Varroa destructor and Apis mellifera to native strains of B. thuringiensis was tested by Alquisira-Ramírez et al. (2014). Also, the effect of different B. thuringiensis strains on the longevity of Africanized honeybee is reported (Libardoni et al. 2018). Pathological observations on midgut (the irregular epithelium profile, numerous irregular cellular fragments of different sizes interspersed with the fibrils of the peritrophic membrane) of adult worker bees under controlled acute exposure to a Bacillus thuringiensis–based biopesticide were observed (D’Urso et al. 2017). Except for the laboratory studies, also the field assessment on the colony performance after aerial application of Bacillus thuringiensis subsp. Kurstaki was done (Lez et al. 2014).
Only one laboratory study reported that surfactin produced by B. subtilis has larvicidal activity against mosquito Aedes aegypti (Revathi et al. 2013).
In addition to the lethal effect of PPPs, very important are their sublethal effects on immune system which protects bees against diseases. Bee immune response is composed of the social (immunity of whole colony) and the individual (immunity of individual bees) immunity. Both parts are linked together (Wilson-Rich et al. 2009). Innate immunity reflects the level of defence against the pathogen infection. Individual immunity consists of the cellular part (the hemocytes) (Hillyer and Strand 2014) and the humoural part (antimicrobial peptides—AMPs, agglutinins, lysines) (Turner 1994). Lysozyme, one from lysines, hydrolyzes the glycosidic bonds in bacterial cell wall. In bees, two types of lysozyme have been identified, the c-type and the i-type (Beckert et al. 2016) with two lysozyme genes encoding the c-type and one gene encoding i-type lysozyme (Evans et al. 2006). On the other hand, AMPs have various activities against bacteria, fungi, and protozoa (Casteels et al. 1993).
Traditionally, the measurement of the acute toxicity of pesticides to beneficial arthropods has relied largely on determination of an acute median lethal dose or concentration. However, the estimated lethal dose during acute toxicity tests may only be a partial measure of the deleterious effects. In addition to direct mortality induced by pesticides, their sublethal effects on arthropod physiology and behaviour should be considered for the complete analysis of their impact (Desneux et al. 2007).
Due to the intensive use of registered PPPs containing B. amyloliquefaciens strain QST 713 in the last decade in many different crops in agriculture, the question has raised whether the use of these products is sufficiently safe for non-target pollinators. We chose winter honeybees as they play a crucial role in overwintering success of colony and according to our assumption described above, they may be exposed to Serenade® ASO containing B. amyloliquefaciens strain QST 713 under realistic field conditions. In our chronic bioassay, we tested direct lethal effect of Serenade® ASO containing B. amyloliquefaciens strain QST 713 as well as the sublethal effects on gut microbiome and immune system.
German national registration report gives us an assumption for slightly higher product application field rate in comparison with that agreed on EU level in 2018. As per hectare, 8 l of Serenade® ASO, corresponding to 8.336 × 1012 CFU, is used, suspended in a total water volume of 200–800 l water. Assuming volume of 200 l as a “worst-case” exposure scenario for non-target pollinators, the concentration of B. amyloliquefaciens strain QST 713 in the spray solution will be 4.17 × 1010 CFU/l or 4.17 × 107 CFU/ml as a maximum application field rate (MAFR) (Anonymous 2014; EFSA 2018).
2 Material and methods
2.1 Chemicals and used doses
In the present study, we used the commercial formulation of Serenade® ASO instead of B. amyloliquefaciens strain QST 713 active ingredient, in order to simulate real agricultural conditions.
Registered PPP named Serenade® ASO contains spores of Bacillus amyloliquefaciens strain QST 713 and several substances produced by this bacterium during fermentation process. It is a biological fungicide formulated as a water dispersible granule, containing a minimum spore concentration of 1 × 1012 colony-forming units (CFU)/kg (equivalent to 13.96 g Bacillus subtilis QST 713/kg product).
2.2 Chronic exposure (cage experiment)
To study the effect of prolonged exposure of Serenade® ASO to winter honeybees, we performed a cage experiment in October 2018 with 10-day chronic exposure scenario according to the proposed method OECD 245 (2017).
All tested colonies, reared in our experimental apiary, were free of obvious symptoms of bee diseases and were located close to the experimental laboratory. No sanitary treatments (e.g. mite or disease treatment measures) were conducted within 3 weeks before the start of the experiment.
Winter adult bees were carefully brushed off from combs and transferred into the laboratory. Test cages in groups of 10 bees/cage were used (five replicates per treatment) and kept in an incubator (33 °C, 55% RH and darkness) for the next 11 days (with 1 day for acclimatisation). Dimethoate as a reference item was used in this experiment.
Because of acclimatisation, all tested bees were fed with a sucrose solution (50% w/w) ad libitum for 24 h prior to exposure. During the exposure phase of next 10 days, caged winter honeybees were offered sugar solutions spiked with nominal concentrations of MAFR of Bacillus amyloliquefaciens QST 713 and 1/10 of MAFR (please see above). Tested winter bees were fed with a minimum spore concentration of 4.17 × 107 CFU/ml diet as the recommended field dose which was confirmed microbiologically on Day 1 and Day 10 of experiment.
Tested bees in the untreated control group were fed a sugar solution without pesticide. Reference item (dimethoate; single nominal concentration of 1.0 mg a.s./kg diet) was used in this bioassay to verify the sensitivity and the reliability of the test system (OECD 245 2017).
The choice of expected dose rate of 40 mg of sugar solution/bee/day was based on our own preliminary experience and on studies published by Schmitzer and Kling (2014) and Decourtye et al. (2003). Caged bees are assumed to share offered sugar solution with applied item due to their social feeding behaviour (trophallaxis). Offered sugar solutions (five replicates per each concentration, positive control, and untreated control) were changed and weighted daily for the 10-day duration of the exposure phase of experiment. Weighing was done before and after feeding to the bees and represents the amount of food consumed by the bees in one cage during 24 h. The amount of weighted food was then divided by the number of living bees at the start of the corresponding feeding interval. Finally, the food consumption was corrected accounting the loss by evaporation (OECD 245 2017).
The positive control and untreated control sugar solutions were prepared 1 day prior to exposure, kept at − 20 °C, and thawed at room temperature before use. Contaminated sugar solutions with tested PPPs were prepared daily fresh. Every 24 h, mortality and behavioural abnormalities were recorded. After 10 days of exposure, winter honeybees were anesthetised by placing the cages in a fridge for 30 min. After this time, bees exposed to the same dose were randomly mixed and transferred to identified plastic containers. A total of 30 tested bees/group (3 × 10 individuals) were analysed by microbiology assay and 10 bees/group were analysed by immunology assay.
2.3 Bacteria isolation and identification
Analysed bees were divided into three main groups: untreated control, MAFR1/1, and MAFR1/10. Every group contained three subsets of ten bees. The intestinal tracts of honeybees (of 10 individuals) were withdrawn under aseptic conditions and homogenised in 1 ml PBS. Serial dilutions were plated over the Nutrient Agar 02-NA02 (Conda, Spain) and incubated for 24 h at 37 °C in thermostat.
The bacterial colonies with different morphology were selected and identified by MALDI-TOF mass spectrometry Microflex LTTM (Bruker Daltonics, Bremen, Germany) instrument using the FlexControlTM version 3.4 software (Bruker Daltonics) or by 16S rDNA analyse.
For this purpose, a part of homogenised digestive tract of bees was used for DNA isolation. The genomic DNA was separated using GenEluteTM Bacterial Genomic DNA Kit (Sigma-Aldrich, USA). The isolated DNA (50 ng) was used as a template in PCR reaction (Taq Core Kit/high yield, Jena Bioscience, Germany). PCR amplification of the 1500 bp eubacterial 16S rDNA was performed using forward (fD1 5′-AGA GTT TGA TCC TGG CTC AG-3′) and reverse (rP2 5′-ACG GCT ACC TTG TTA CGA CTT-3′) primers (Weisburg et al. 1991). The PCR products were separated by electrophoresis (PowerPac Basic, Bio-Rad, USA) in 1% agarose gel (Sigma-Aldrich) in TAE buffer (Sambrook et al. 1989).
The amplified product was purified from agarose gel using Wizard SV Gel and PCR Clean-Up System (Promega, Madison, USA) and cloned using InsTAcloneTM PCR Cloning Kit (ThermoFisher Scientific, Colorado, USA). Recombinant plasmids were selected on Luria–Bertani Agar (Becton & Dickinson, USA) plates with ampicillin and kanamycin addition. The selected recombinant plasmids were isolated by GenElute Plasmid MiniPrep Kit (Sigma-Aldrich) and sequenced by the dideoxytermination method using universal M13 sequencing primers at Eurofins Genomics (Germany). The obtained sequences were subjected to BLAST search against GenBank database.
2.4 B. amyloliquefaciens QST 713 antibacterial activity
The effect of overnight culture of B. amyloliquefaciens QST 713 cell-free supernatant on the other bacteria isolated from the digestive tract of tested bees was investigated. The cell-free supernatant (CFS) was prepared as follows: single cell colony of B. amyloliquefaciens QST 713 was inoculated to Tryptic Soy Broth-TSB (Conda, Spain) and incubated at 37 °C overnight. The CFS was obtained by centrifugation (14,000 rpm for 2 min at laboratory temperature) and was filtered through a bacteriological filter to eliminate the present bacteria. For testing the potential antimicrobial activity of B. amyloliquefaciens QST 713 against other bacterial isolates from the gastrointestinal tract of honeybees, the disc diffusion method was used (Balouiri et al. 2016). The Nutrient Agar 02 plate surface was inoculated by 100 μl of 0.5 McFarland tested bacterial suspensions. Subsequently, sterile filter paper discs were placed on the agar surface and then the discs were impregnated with 20 μl CFS of B. amyloliquefaciens QST 713. After incubation at 37 °C overnight, the potential inhibition growth zones were observed.
2.5 Isolation of DNA and PCR amplification
The total DNA from fresh intestinal samples of honeybees for DGGE analysis was extracted using NucleoSpin DNA Stool (MACHEREY-NAGEL GmbH Co.KG, Germany). Isolated DNA was used as a template for PCR amplification of 16S rRNA gene fragments with used specific bacterial primers GC-clamp-968f (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAA CGC GAA GAA CCT TAC-3′) and 1401r (5′-CGG TGT GTA CAA GAC CC-3′) (Nübel et al. 1996). All PCR reactions were performed in a 50-μl PCR mixture containing 1 μl of the substrate of DNA, 1 × PCR buffer, 2 mmol/l MgCl2, 1 μl of a 200 μmol/l of each dNTP, 1.25 U Platinum Taq DNA polymerase (Invitrogen, CA USA), and 25 pmol each primer using C1000TM Thermal Cycler (Bio-Rad Laboratories, USA). The cycling conditions were 94 °C for 5 min, 9 cycles of 94 °C for 1 min, 45 °C for 1 min, 72 °C for 1 min; 14 cycles of 94 °C for 1 min, 60 °C for 1 min, 72 °C for 1 min followed by final extension at 72 °C for 10 min. PCR products were detected using 1% agarose gel electrophoresis containing ethidium bromide and photographed using Gel Logic 212 PRO Imaging system (Carestream, NY, USA). PCR products generated with GC-clamp-968f and 1401r primers were subjected to DGGE analysis.
2.6 Denaturing gradient gel electrophoresis
DGGE was performed using DCodeTM Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, CA, USA). The round II PCR reaction products in a total volume of 45 μl were loaded onto 8% (w/v) polyacrylamide gel (40% Acrylamide-Bis 37.5:1) in 1 × TAE (40 mM Tris, 20 mM acetate, 1 mM EDTA) containing a linear denaturing gradient ranging from 30–60% denaturant (100% denaturant solution consists of 7 M urea and 40% formamide). Electrophoresis was run for 17 h at a constant voltage of 50 V and a temperature of 60 °C. After electrophoresis, the gel was incubated for 20 min in ethidium bromide (0.5 μg/ml), rinsed for 20 min in distilled water, and photographed with UV transillumination using a Fusion Fx7 The Multi-Application System (Adelab Scientific, Thebarton SA).
2.7 Isolation of RNA and cDNA synthesis for testing the immunomodulation effect
The intestinal tracts of anesthetised bees (+ 4 °C for 10 min) were withdrawn under aseptic conditions (n = 10/group). Received guts were washed with PBS. The total RNA of these guts was extracted using Purezol™ reagent (Bio-Rad, Hercules, CA) following the manufacturer’s instructions. For removal of genomic DNA from RNA samples, we used DNA Rapid Removal Kit (Thermo Scientific, Waltham, MA, USA). RNA purity and concentration were determined spectrophotometrically at 260/280 nm by Nanodrop 8000 (Thermo Scientific, Waltham, MA). Total RNA was immediately reverse transcribed by RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). Resulting cDNA was used as a template in qPCR.
2.8 Gene expression analysis (qPCR)
We determined gene expression of Abaecin, Apidaecin, Defensin-1, Hymenoptaecin, Lysozyme-2, and reference β-actin. Primers for gene expression are listed in Table I. For amplification and detection of specific products, we used iCycler CFX96 (Bio-Rad, Hercules, CA). Ten microlitres reaction volume contained iQ™ SYBR® Green Supermix (Bio-Rad, Hercules, CA), 0.5 μM of each primer, and 40 ng/μL of cDNA. All the reactions were performed in triplicate and negative control without cDNA template. The experimental protocol was composed of the initial denaturation 95 °C for 5 min, 35 cycles of 3 steps: 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s, and final extension at 72 °C for 5 min was followed by melting curve analysis. A melting curve from 59 to 92 °C with reading at every 0.5 °C was performed for each individual real-time PCR plate. Reaction efficiency (100 ± 5%) was calculated for each assay. Relative normalised expression was calculated by the 2−ΔΔCT method using CFX96Manager Software (Bio-Rad, Hercules, CA).
2.9 Statistical analysis
The statistical processing of the obtained data was performed by using ToxRat Professional® (version 3.2.1; ToxRat Solutions GmbH). Fisher’s Exact Test (Bonferroni-Holms corrected, one-sided, p ≤ 0.05) was used to evaluate whether there are significant differences between the mortality data of both the Serenade® ASO treatment groups and untreated control group and to determine the NOEC based on observed mortality.
Experimental results of gene expression were performed in triplicate and expressed as mean ± standard deviation (SD). Data were evaluated using the GraphPad Prism 3.00 software by one-way analysis of variance (ANOVA) followed by Tukey’s Multiple Comparison Test.
3 Results
3.1 Chronic exposure (cage experiment)
We investigated the lethal and sublethal effects of chronic exposure of commercial PPP containing Bacillus amyloliquefaciens QST 713 spores to laboratory-reared winter adult honeybees (OECD 245 2017). Tested bees were fed daily with doses corresponding to the maximum product application field rate (1/1 MAFR) and its 1/10. Table II (below) summarises applied and consumed dosages (nominal and measured) over 10-day chronic test. The theoretical daily food consumption of 40.0 mg diet/bee/day was slightly exceeded except for the positive control group, in which overall mean daily syrup intake of 21.4 ± 2.2 mg/bee/day was observed. The number of applied CFUs of the strain QST 713 was confirmed microbiologically on Day 1 and Day 10, in both tested groups which were close to the expected nominal values (Table II).
Under our laboratory conditions, 1/1 MAFR and 1/10 MAFR did not cause dose-dependent mortality. Observed mortality in untreated control group over the entire test period was 12% compared with 100% in the positive control group observed on Day 8 (see Table III). No behavioural abnormalities were observed throughout the entire exposure period of 10 days in all treatment groups, except for the positive control group (dimethoate), in which six bees were categorised as affected (hyperactivity, aggressiveness, increased self-cleaning behaviour) at Day 6 of experiment.
Fisher’s exact test (Bonferroni-Holms corrected, one-sided, p ≤ 0.05) showed that the NOEC appears to be higher than or equal to 4.17 × 107 CFU/ml diet, resp. 4.17 × 1010 CFU/l diet.
3.2 Microbiological and immunological screening
The Serenade® ASO containing spores of B. amyloliquefaciens QST 713 was used also for testing the potential effect of this fungicide on the microbial community of digestive tracts of tested winter adult bees by direct microbial analysis or by DGGE.
The untreated control groups of tested bees were recorded on the average of 4.43 × 103 CFU/ml of bacteria. After 10 days of feeding period with Bacillus amyloliquefaciens, QST 713 in MAFR 1/1 group was detected 6.06 × 106 CFU/ml and in the MAFR 1/10 group was counted 8.56 × 105 CFU/ml of bacteria respectively. From untreated control samples, we have isolated and sequenced overall 11 bacterial clones. The analysis of the 16S rDNA sequences showed that 6 of them are grouped in the genus Bacillus. Other bacteria were identified as Morganella sp., Hafnia sp., Staphylococcus sp., Brevibacterium sp., and Kosakonia sp. We have tested the potential influence of CFS of overnight B. amyloliquefaciens QST 713 culture on the growth of all cultivable bacterial species isolated from untreated control trials. Practically, only two of the tested bacterial species, Bacillus sp. and Brevibacterium sp., were lightly sensitive to the CFS of B. amyloliquefaciens QST 713 (Figure 1), which suggests that tested fungicide should not have a significant effect on the biodiversity of bee digestive tract. The DGGE analysis also confirmed no significant changes in the composition of bacterial community within two different treatments by Serenade® ASO after 10 days of chronic test (Figure 2).
DGGE analysis of total DNA isolated from whole intestinal tracts of winter adult bees (3 × 10/3) treated or not with commercial formulation of Serenade® ASO containing spores of B. amyloliquefaciens strain QST 713 over 10 days chronic test. Lane 1, 2, 3—untreated controls; Lane 4, 5, 6—samples treated with Serenade® ASO-MAFR/1; Lane 7, 8, 9—samples treated with Serenade® ASO-MAFR/10. Lane 10—total of DNA of B. amyloliquefaciens QST 713.
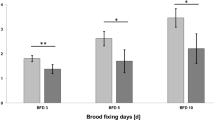
In addition to the previous tests, we investigated the effect of plant protection product containing spores Bacillus amyloliquefaciens QST 713 on bee immune system. We focused on gene expression encoding antimicrobial compounds. Relative gene expression of real-time PCR was used to study the expression level for each antimicrobial compound (Figure 3). Gene expression of Abaecin was on the same level in the treated groups but lower in comparison with the untreated control group. The same trend of gene expression was noted for Apidaecin; however, lower gene expression was statistically significant (P < 0.01) in both groups in comparison with untreated control. Gene expression of Defensin-1, Hymenoptaecin, and Lysozyme-2 was lower in the 1/1 MAFR group in comparison with the 1/10 MAFR group. Significantly lower gene expression was recorded in Defensin-1 in the 1/1 MAFR group (P < 0.05) and in the case of Hymenoptaecin in both experimental groups (P < 0.01) as compared with control. Abaecin and Lysozyme-2 gene expression did not show significant differences due to higher standard deviations.
4 Discussion
The risk of commercial PPP containing B. amyloliquefaciens QST 713 to honeybees cannot be excluded as application can potentially result in their exposure either through direct over-spray or by contact with “spores” on plants, while bees are foraging for food.
In our in vitro bioassay, caged winter honeybees were fed 10 days with a sucrose solution (50% w/w) ad libitum spiked with PPP at maximum application field rate of 4.17 × 107 CFU/ml (EFSA 2018). According to Biopesticide registration action document on Bacillus subtilis strain QST 713, there were no behavioural or morphological abnormalities seen in any of the treatments in the two dietary whole hive studies conducted until total adult honeybee emergence (22 and 24 days) (EPA 2006).
The theoretical food consumption of 40.0 mg diet/bee/day was reached in almost all the tested groups, except the positive control group with overall mean syrup intake of 21.4 ± 2.2 mg/bee/day (dimethoate). Nevertheless, to that fact, clear toxic effects in this group were seen giving an assumption that the test system with adult winter bees was sensitive.
Observed mortality in both, 1/1 MAFR and 1/10 MAFR test groups, did not cause a dose-dependent mortality. Mortality observed in the untreated control group over the entire test period was 12% compared with 100% in the positive control group observed on Day 8 (see Table II), thus making this bioassay valid (OECD 245 2017).
Natural variations in the honeybee gut microbial populations are affected by multiple factors, e.g. ontogenetic stage, age, and geographic location. Bacterial counts in the gastrointestinal tract of honeybees show clear yearly cycles (summer–winter) within a colony but vary on the individual level as well (Hroncova et al. 2015).
The fingerprinting techniques are a rapid and very easy alternative to the analysis of microbial communities. DGGE is a fingerprinting technique commonly used to study genetic diversity of the DNA genes coding for ribosomal RNA. The 16S rRNA genes are used for phylogenetic affiliation of Eubacteria and Archaea. The uniqueness of this technique is that DNA fragments of the same size but with differing base pair sequences can be separated (Muyzer et al. 1993). The DGGE techniques have been used in environmental microbiology, in the study of variation, the honeybee gut microbial diversity, food microbiology etc. (Gafan et al. 2005; Hroncova et al. 2015).
In our experiments, we used the DGGE analysis for the study of influence commercial formulation of Serenade® ASO containing spores of B. amyloliquefaciens strain QST 713 on the microbial community of winter adult bees. From the experimental groups, containing predominantly Bacillus amyloliquefaciens QST 713, the natural bee bacterial isolates were hardly recovered. In general, the bacterial counts in the digestive tracts of experimental winter bees in the untreated control group were relatively low, only 4.43 × 103 CFU/ml. Only two of the isolated bacterial species Bacillus sp. (graminis) and Brevibacterium sp. were lightly sensitive to the CFS of B. amyloliquefaciens QST 713. Traditional cultivation methods used for isolation and identification of microbial communities identify only a small fraction (from 0.01–10%) of the total microbial biomass (Torsvik el al. 1990; Ward et al. 1990). On the other hand, to create a visible band on the DGGE gel, a bacterial species has to constitute approximately 1% of the entire population (Casamayor et al. 2000). The results of our DGGE analysis showed no significant changes in the composition of bacterial community between the untreated control and experimental groups after 10 days of chronic exposure by commercial formulation Serenade® ASO.
As in many animals, honeybees rely on their gut microbial community for a variety of functions, including food processing, regulation of immune system, and defence against pathogens. Perturbations of this system have the potential to lead to negative consequences for host fitness (Motta et al. 2018). Healthy endogenous gut microbiota can stimulate immune system in bees, like in other animals (Wu et al. 2013). Lactic acid bacteria have a positive effect on the immune system of bees through higher synthetisation of antimicrobial peptides (Evans and Lopez 2004). Whereas DGGE analysis did not show significant changes in bees gut microbiota, we suppose that gene level of antimicrobial compounds is affected above all by the influence of Serenade® ASO containing spores of B. amyloliquefaciens.
Many studies have investigated effect of pesticides on bee colonies with a negative impact on immunity (Christen and Fent 2017; Aufauvre et al. 2014; Garrido et al. 2013; Di Prisco et al. 2013). Motta et al. (2018) described the potential mechanism by which glyphosate affects bee health. While some species in the bee gut can tolerate high concentrations of glyphosate due to the presence of a class II EPSPS enzyme, others are sensitive due to the presence of a class I EPSPS.
Specifically, the influence of PPPs on the immune system is very important since it is responsible for the elimination of pathogens. AMPs are mainly part of innate immunity. Bee fat body produces AMPs and excretes them to the open circulatory system where they attack bacterial pathogens (Wang et al. 2014). However, epithelial cells of gut can also synthetize AMPs (Tsakas and Marmaras 2010). The mechanism of action is the creation of leaks into prokaryotic membranes or inhibition of translation or folding of bacterial protein. The expression of genes encoding AMPs is regulated by Imd/JNK and/or Toll signalling pathways (Danihlík et al. 2015). These pathways start after recognition of peptidoglycans via Peptidoglycan Recognition Proteins (PGRPs) (Brutscher et al. 2017).
In this study, two groups of bees were fed with 1/1 MAFR and 1/10 MAFR of Serenade® ASO containing spores of B. amyloliquefaciens strain QST 713 and the untreated control group received sugar solution without the pesticide. The expression of genes encoding antimicrobial peptides was decreased in one/both treated groups in comparison with the untreated control group. Significantly lower expression was observed in genes for Apidaecin, Defensin-1, and Hymenoptaecin, thus indicating an immunosuppressive effect of tested pesticide.
Yan et al. (2016) described host immunosuppression by antimicrobial compounds macrolactins derivatives produced by B. subtilis B5. Another molecule Surfactin produced by B. subtilis has potential immunosuppressive effect through inhibition of NF-κB and other immunologically important molecules (Park and Kim 2009). Bees have similar signalling pathways as mammalians, bee Dorsal molecule for Toll signalling pathway, and Relish molecule for Imd signalling pathway are homologues of mammalian NF-κB (Lourenço et al. 2018).
We hypothesise that antimicrobial products of B. amyloliquefaciens QST 713 could partly downregulate gene expression of bee AMPs and subsequently reduce the immune response to combat the potential pathogens, as well as they may potentially affect the physiological microflora (but this needs to be reflected in future research).
5 Conclusion
Pesticide risk assessment scheme used in re-registration process all over the world uses the determination of the acute/chronic toxicity of pesticides to beneficial arthropods, bees included.
Results of this study suggest that more attention should be paid to the sublethal effects of commonly used commercial plant protection products containing microbial active substances. In conclusion, no treatment significantly affected the survival of caged adult winter bees or their eubacterial microflora after 10 days of exposure to commercial product Serenade® ASO containing spores of Bacillus amyloliquefaciens QST 713 (formerly subtilis).
However, clear statistically significant decrease in some humoural parameters of bee immune system was detected in one/both tested treatments.
As the social immunity of bee colony is more complex and depends on immune response of individual bees as well, any sublethal adverse effect of the PPPs and their mixtures on individual basis may finally contribute to weakening of the whole bee colony. This subsequently may have negative consequences for agriculture and food production. The sublethal effect of commercial formulations containing microorganisms on individual and social bee level needs to be further studied.
References
Alquisira-Ramírez, E.V., Paredes-Gonzalez, J.R., Hernández-Velázquez, V.M., Ramírez-Trujillo, J.A., Pe˜na-Chora, G. (2014) In vitro susceptibility of Varroa destructor and Apismellifera to native strains of Bacillus thuringiensis. Apidologie 45, 707–718.
Anonymous (2014) National assessment report [Internet]. [cited 2019 September 3] Available at: https://www.bvl.bund.de/SharedDocs/Downloads/04_Pflanzenschutzmittel/01_zulassungsberichte/007918-00-00.html?nn=1406458
Aufauvre, J., Misme-Aucouturier, B., Viguès, B., Texier, C., Delbac, F., Blot, N. (2014) Transcriptome Analyses of the Honeybee Response to Nosema ceranae and Insecticides. PLoS ONE 9(3): e91686.
Balouiri, M., Sadiki, M., Ibnsouda, S.K. (2016) Methods for in vitro evaluating antimicrobial activity: A review. Journal of Pharmaceutical Analysis, 6:71-79.
Beckert, A., Wiesner, J., Schmidtberg, H., Lehmann, R., Baumann, A., Vogel, H., Vilcinskas, A. (2016) Expression and characterization of a recombinant i-type lysozyme from the harlequin ladybird beetle Harmonia axyridis. Insect Molecular Biology, 25, 202-215.
Boncristiani, H., Robyn Underwood, R., Ryan Schwarz, R., Evans, D. J., Pettis, J., van Engelsdorp, D. (2012) Direct effect of acaricides on pathogen loads and gene expression levels in honey bees Apis mellifera. In Journal of Insect Physiology 58, 613–620
Brutscher, L.M., Daughenbaugh, K.F., Flenniken, M.L., (2017) Virus and dsRNA-triggered transcriptional responses reveal key components of honey bee antiviral defense. In Scientific Reports 7: 6448
Casamayor, O., Schäfer, H., Baneras, L., Pedros-Alio, C. and Muyzer, G. (2000) Identification of and spatio-temporal differences between microbial assemblages from two neighbouring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499–508.
Casteels, P., Ampe, C., Jacobs, F., Tempst, P. (1993) Functional and chemical characterization of hymenoptaecin, an antibacterial polypeptide that is infection-inducible in the honey bee (Apis mellifera). Journal of Biological Chemistry, 268, 7044–7054.
Christen, V. and Fent, K. (2017) Exposure of honey bees (Apis mellifera) to different classes of insecticides exhibit distinct molecular effect patterns at concentrations that mimic environmental contamination. Environmental Pollution. Volume 226, 48-59
Cizelj, I., Glavan, G., Božič, J., Oven, I., Mrak, V., Narat, M. (2016) Prochloraz and coumaphos induce different gene expression patterns in three developmental stages of the Carniolan honey bee (Apis mellifera carnica Pollmann). Pesticide Biochemistry and Physiology, 128, 68–75.
Danihlík, J., Aronstein, K., Petřivalský, M. (2015) Antimicrobial peptides: a key component of honey bee innate immunity: Physiology, biochemistry, and chemical ecology. Journal of Apicultural Research 54(2), 123-136.
Decourtye, A., Lacassie, E., Pham-Delégue, M. (2003) Learning performances of honeybees (Apis mellifera L.) are differentially affected by imidacloprid according to the season. Pest. Manag. Sci. 59, 269-278.
Desneux, N., Decourtye, A., Delpuech, J.M., 2007. The sublethal effects of pesticides on beneficial arthropods. Annu. Rev. Entomol. 52, 81-106.
Di Prisco, G., Cavaliere, V., Annoscia, D., Varricchio, P., Caprio, E., Nazzi, F., Gargiulo, G., Pennacchio, F. (2013) Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. PNAS 110 (46) 18466-18471.
D’Urso, V., Mazzeo, G., Vaccalluzzo, V., Sabella, G., Bucchieri, F., Viscuso, R., Vitale, D.G.M. (2017). Observations on midgut of Apis mellifera workers (Hymenoptera:Apoidea) under controlled acute exposures to a Bacillus thuringiensis-based biopesticide. Apidologie 48, 51–62.
EFSA (2018) Renewal Assessment Report Bacillus amyloliquefaciens strain QST 713 Serenade ASO Volume 3 – B.9 Effects on non-target organisms [Internet]. [cited 2019 May 14] Available at: https://www.efsa.europa.eu/en/consultations/call/180905
EPA (2006) Bacillus subtilis Strain QST 713 (006479) Biopesticide Registration Action Document. [Internet]. [cited 2019 May 14] Available at: https://www.researchgate.net/profile/Houda_Kawas/post/How_can_I_evaluate_the_effect_of_pesticides_on_the_growth_of_Bacillus_subtitles_biological_agent/attachment/5ab3ddddb53d2f0bba59a900/AS%3A607023888605191%401521737181639/download/Bacillus+subtilis+Strain+QST+713.pdf
Evans, J.D., Aronstein, K., Chen, Y.P., Hetru, C., Imler, J.L., Jiang, H., Kanost, M., Thompson, G.J., Zou, Z., Hultmark, D. (2006) Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Molecular Biology, 5, 645–656.
Evans, J.D and Lopez, M.L. (2004) Bacterial Probiotics Induce an Immune Response in the Honey Bee (Hymenoptera: Apidae). J. Econ. Entomol. 97(3): 752-756.
Frabotta, D. (2014) Stricter MRLs: pushing global demand for biopesticides. Farm Chemicals International, v.28, n.28, p.18
Gafan, G.P., Lucas, V.S., Roberts, G.J., Petrie, A., Wilson, M. and Spratt, D.A. (2005) Statistical analyses of complex denaturing gradient gel electrophoresis profiles. J Clin Microbiol 43:3971–3978
Garrido, P.M., Antúnez, K., Martín, M., Porrini, M.P., Zunino, P., Eguaras, M.J., (2013) Immune-related gene expression in nurse honey bees (Apis mellifera) exposed to synthetic acaricides. In Journal of Insect Physiology 59(1) 113-119.
Hillyer, J.F. and Strand, M.R., 2014. Mosquito hemocyte-mediated immune responses. Curr Opin Insect Sci 3, 14-21.
Hroncova, Z., Havlik, J., Killer, J., Doskocil, I., Tyl, J, Kamler, M., Titera, D., Hakl, J., Mrazek, J., Bunesova, V. and Rada, V. (2015) Variation in Honey Bee Gut Microbial Diversity Affected by Ontogenetic Stage, Age and Geographic Location. PLoS ONE 10, e0118707
Khongphinitbunjong, K., De Guzman, L. I., Tarver, M. R., Rinderer, T. E., Chen, Y., Chantawannakul, P. (2015) Differential viral levels and immune gene expression in three stocks of Apis mellifera induced by different numbers of Varroa destructor. Journal of Insect Physiology, 72, 28–34.
Lez, M.D.M., Llado, G., Petro, A.B., Alemany, A. (2014) First field assessment of Bacillus thuringiensis subsp. Kurstaki aerial application on the colony performance of Apis mellifera L. (Hymenoptera: Apidae). Spanish J. Agric. Res. 12, 405–408.
Libardoni, G., De Gouvea, A., Costa Maia, F., Lozano, E. Freitas, P., Caroline Colombo, F., Raulino, F., Mendes AntunesMaciel, R., Potrich, M. (2018) Effect of different Bacillus thuringiensis strains on the longevity of Africanized honey bee. Semina: Ciências Agrárias. 39. 329. https://doi.org/10.5433/1679-0359.2018v39n1p329.
Lourenço, P., Florecki, M.M., Simões, Z.L.P., Evans, J. D. (2018) Silencing of Apis mellifera dorsal genes reveals their role in expression of the antimicrobial peptide defensin-1. Insect Molecular Biology 27(5), 577–589.
Motta, E. V. S., Raymanna, K. and Morana N. A. (2018) Glyphosate perturbs the gut microbiota of honey bees. Proc Natl Acad Sci USA. 2018 Oct 9; 115(41):10305–10.
Muyzer, G., de Waal, E. C., Uitterlinden, A. G. (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700
Nübel, U., Engelen, B., Felske, A., Snaidr, J., Wieshuber, A., Amann, R.I. (1996) Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol 178: 5636–5643.
Olson, S. (2015) An analysis of the biopesticide market now and where it is going. Outlooks on Pest Management 26:203–206
OECD (2017) Guideline for the Testing of Chemicals No. 245: Honey bee (Apis mellifera l.), chronic oral toxicity test (10-day feeding), [Internet]. [cited 2019 May 14] Available at: https://www.oecd-ilibrary.org/docserver/9789264284081-en.pdf?expires=1557818342&id=id&accname=guest&checksum=1DFD4BB165B9F393A76B1713A732E9DA
Park, S.K. and Kim, Y.H. (2009) Surfactin inhibits immunostimulatory function of macrophages through blocking NK-κB, MAPK and Akt pathway. International Immunopharmacology 9(7–8), 886-893.
Revathi, K., Chandrasekaran, R., Thanigaivel, A., Kirubakaran, S.A., SathishNarayanan, S., Senthil-Nathan, S. (2013) Effects of Bacillus subtilis metabolites on larval Aedes aegypti L. Pestic. Biochem. Physiol. 107, 369–376.
Sambrook, J., Fritsch, E.F., Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. 2nd Ed. New York, Cold Spring Harbor Laboratory Press, Cold Spring Harbor.
SANCO (2006) Review report for the active substance Bacillus subtilis QST 713 [Internet]. [cited 2019 September 3] Available at: https://ec.europa.eu/food/plant/pesticides/eu-pesticides-database/public/?event=activesubstance.detail&language=EN&selectedID=986
Schmitzer S. and Kling A. (2014) Final Report – Summary of the Results of the International Ring Test for the Standardisation of a 10 Day Chronic Feeding Test on Honey Bees (Apis mellifera L.) in the Laboratory. [Internet]. [cited 2019 May 14] Available at: https://www.oecd.org/env/ehs/testing/Final_Report_of_Ringtest_Group_Bee_Chronic_Lab_Test_2014%20(2).pdf
Torsvik V., Goksoyr J., Daae F.L. (1990) High diversity in DNA of soil bacteria. Applied and Environmental Microbiology, 56: 782–787.
Tsakas S. and Marmaras V.J. (2010) Insect immunity and its signalling: an overview. Invertebr. Survival 7: 228-238.
Turner, R.J. Immunology – A comparative Approach. 1994. 222p. ISBN ISBN 10: 0471944009 / ISBN 13: 9780471944003
Wang, J.L., Chen, L., Tang, L., Zhao, H.B., Liu, X.S., Wang, Y.F. 2014. 20-hydroxyecdysone transcriptionally regulates humoural immunity in the fat body of Helicoverpaarmigera. In Insect Mol. Biol 23, 842-856.
Ward, D.M., Weller, R., Bateson, M.M. (1990) 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 3; 345(6270):63-5.
Weisburg, W.G., Barns, S.M., Pelletier, D.A., Lane, D.J. (1991) 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology, 173:697-703.
Wilson-Rich, N., Spivak, M., Feffetman, N.H., Starks, P.T. (2009) Genetic, individual, and group facilitation of disease resistance in insect societies. In Annu Rev Entomol 54, 405-423.
Wu, M., Y. Sugimura, Taylor, D., Yoshiyama, M. (2013) Honeybee gastrointestinal bacteria for novel and sustainable disease control strategies. J. Develop. Sustain. Agr. 8: 85–90.
Yan, X., Zhou, Y.X., Tang, X.X., Liu, X.X., Yi, Z.W., Fang, M.J., Wu, Z., Jiang, F.Q., Qiu, Y.K. (2016) Macrolactins from Marine-Derived Bacillus subtilis B5 Bacteria as Inhibitors of Inducible Nitric Oxide and Cytokines Expression. Mar. Drugs 14, 195.
Funding
Funding was provided by the Slovak Grant Agency VEGA (grant no. 2/0100/19) and by the National Reference Laboratory for Pesticides of University of Veterinary Medicine and Pharmacy in Košice, Slovakia.
Author information
Authors and Affiliations
Contributions
All authors have contributed equally to the work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Manuscript editor: Monique Gauthier
Effets sublétaux sur les abeilles adultes d’hiver par des produits commerciaux phytosanitaires contenant des sporesBacillus amyloliquefaciens QST 713 (anciennementsubtilis).
Apis mellifera / Bacillus amyloliquefaciensQST713/ exposition / toxicité / microbiome intestinal / système immunitaire.
Sublethale Effekte bei adulten Winterbienen durch kommerzielle Pflanzenschutzprodukte, die Sporen von Bacillus amyloliquefaciens QST 713 (früher subtilis ) enthalten.
Apis mellifera / Bacillus amyloliquefaciens QST 713 / Exposition / Toxizität / Darmmikrobiom / Immunsystem.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sabo, R., Kopčáková, A., Hamarová, Ľ. et al. Sublethal effects of commercial plant protection product containing spores Bacillus amyloliquefaciens QST 713 (formerly subtilis) on winter adult honeybees. Apidologie 51, 226–239 (2020). https://doi.org/10.1007/s13592-019-00705-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-019-00705-9